6VWW
 
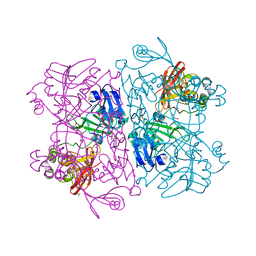 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | 分子名称: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-20 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6W61
 
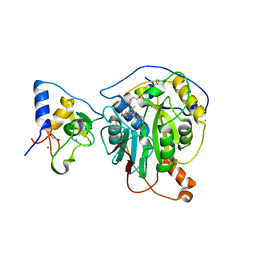 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
6W01
 
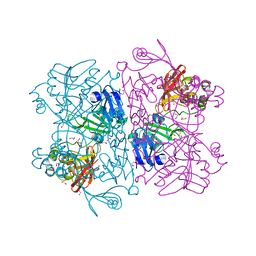 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-28 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6WTC
 
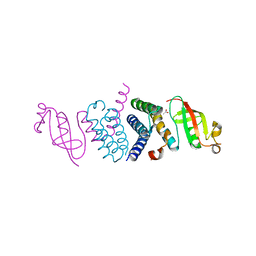 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-02 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
3PMD
 
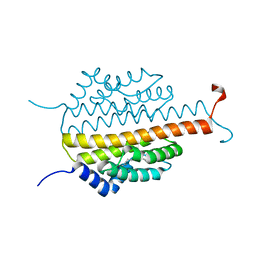 | | Crystal structure of the sporulation inhibitor pXO1-118 from Bacillus anthracis | | 分子名称: | CHLORIDE ION, Conserved domain protein, UNDECANOIC ACID | | 著者 | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | 登録日 | 2010-11-16 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
3PMC
 
 | | Crystal structure of the sporulation inhibitor pXO2-61 from Bacillus anthracis | | 分子名称: | CHLORIDE ION, IODIDE ION, Uncharacterized protein pXO2-61/BXB0075/GBAA_pXO2_0075 | | 著者 | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | 登録日 | 2010-11-16 | | 公開日 | 2011-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
1MG7
 
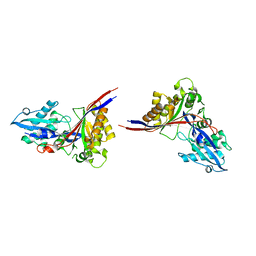 | | Crystal Structure of xol-1 | | 分子名称: | early switch protein xol-1 2.2k splice form | | 著者 | Luz, J.G, Hassig, C.A, Godzik, A, Meyer, B.J, Wilson, I.A. | | 登録日 | 2002-08-14 | | 公開日 | 2003-04-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators
Genes Dev., 17, 2003
|
|
6W4H
 
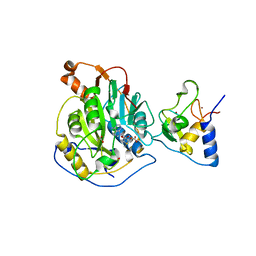 | | 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 | | 分子名称: | 2'-O-methyltransferase, ACETATE ION, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6W75
 
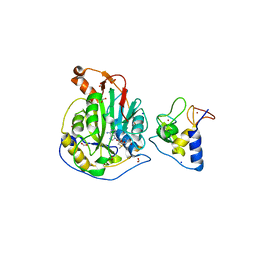 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | 分子名称: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6DFP
 
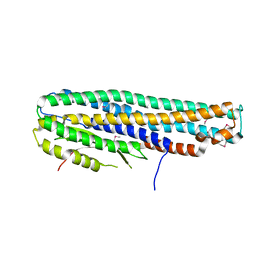 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | 分子名称: | VCA0883 | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-15 | | 公開日 | 2018-05-23 | | 最終更新日 | 2022-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6OV8
 
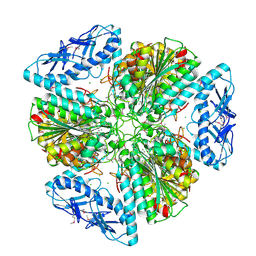 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-07 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
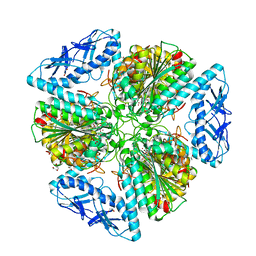 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | 分子名称: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-03-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
5CAG
 
 | |
5CXT
 
 | |
3U21
 
 | |
4DGU
 
 | |
4EPS
 
 | |
3GF8
 
 | |
1J5S
 
 | |
1J6U
 
 | |
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | 著者 | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2022-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
1KQ3
 
 | | CRYSTAL STRUCTURE OF A GLYCEROL DEHYDROGENASE (TM0423) FROM THERMOTOGA MARITIMA AT 1.5 A RESOLUTION | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ZINC ION, ... | | 著者 | Wilson, I.A, Miller, M.D, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2002-01-03 | | 公開日 | 2002-02-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQ4
 
 | |
