3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFD
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AEK
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AEM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
4AFM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens. | | Descriptor: | ACETATE ION, ENDOGLUCANASE CEL5A, GLYCEROL | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AZZ
 
 | |
4AYQ
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYP
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside | | Descriptor: | CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, SODIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4B1L
 
 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AYO
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4B1M
 
 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AYR
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4CD8
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD5
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD7
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG and beta-1,4-mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4C91
 
 | | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility | | Descriptor: | ALPHA-GLUCURONIDASE GH115, CITRATE ANION, D-glucuronic acid | | Authors: | Rogowski, A, Basle, A, Farinas, C.S, Solovyova, A, Mortimer, J.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Evidence that Gh115 Alpha-Glucuronidase Activity is Dependent on Conformational Flexibility
J.Biol.Chem., 289, 2014
|
|
4C1S
 
 | | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 76 MANNOSIDASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4CD4
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4C1R
 
 | | Bacteroides thetaiotaomicron VPI-5482 mannosyl-6-phosphatase Bt3783 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MANNOSYL-6-PHOSPHATASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4CD6
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4C90
 
 | | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility | | Descriptor: | ALPHA-GLUCURONIDASE GH115, SODIUM ION | | Authors: | Rogowski, A, Basle, A, Farinas, C.S, Solovyova, A, Mortimer, J.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evidence that Gh115 Alpha-Glucuronidase Activity is Dependent on Conformational Flexibility
J.Biol.Chem., 289, 2014
|
|
4D4B
 
 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4C
 
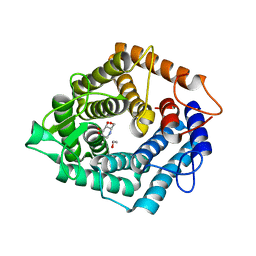 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManDMJ | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4D
 
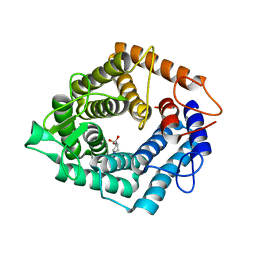 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManIFG | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4A
 
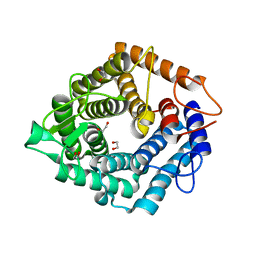 | | Structure of the catalytic domain (BcGH76) of the Bacillus circulans GH76 alpha mannanase, Aman6. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
