1KV8
 
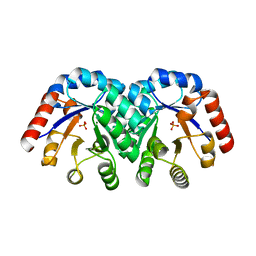 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
3JZU
 
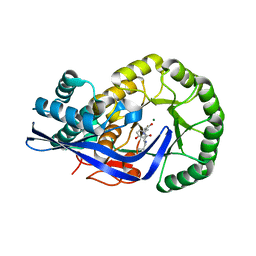 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1TKK
 
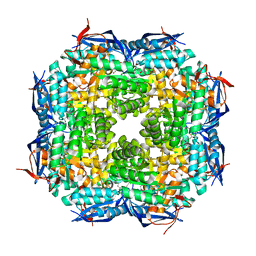 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
2QJN
 
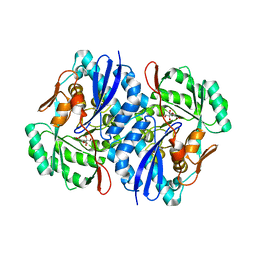 | | Crystal structure of D-mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and 2-keto-3-deoxy-D-gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
1TEL
 
 | | Crystal structure of a RubisCO-like protein from Chlorobium tepidum | | Descriptor: | ribulose bisphosphate carboxylase, large subunit | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a RubisCO-like protein from Chlorobium tepidum
To be Published
|
|
2QJM
 
 | | Crystal structure of the K271E mutant of Mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and D-mannonate | | Descriptor: | D-MANNONIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
3BOX
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg | | Descriptor: | L-rhamnonate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
3CB3
 
 | | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate | | Descriptor: | L-GLUCARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate.
To be Published
|
|
4DO7
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | Descriptor: | Amidohydrolase 2, SULFATE ION, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
4DF0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus
To be Published
|
|
4DFD
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
3SJ3
 
 | | Crystal structure of the mutant R160A.Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3SJN
 
 | | Crystal structure of enolase Spea_3858 (target EFI-500646) from Shewanella pealeana with magnesium bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Spea_3858 from Shewanella Pealeana
To be Published
|
|
3SBF
 
 | | Crystal structure of the mutant P311A of enolase superfamily member from VIBRIONALES BACTERIUM complexed with Mg and D-Arabinonate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-arabinonic acid, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the mutant P311A of enolase superfamily member from Vibrionales bacterium complexed with Mg and D-Arabinonate
To be Published
|
|
3SIZ
 
 | | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4LFE
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, partially liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl diphosphate synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LLS
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP, and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP and calcium bound in active site
To be Published
|
|
4LFG
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, fully liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl Diphosphate Synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LLT
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, Geranyltranstransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site
To be Published
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | Descriptor: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4LOB
 
 | | Crystal structure of polyprenyl diphosphate synthase A1S_2732 (Target EFI-509223) from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, GLYCEROL, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of polyprenyl diphosphate synthase from Acinetobacter baumannii
To be Published
|
|
4WK5
 
 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
4WT7
 
 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
