2H6R
 
 | | Crystal Structure of triosephosphate isomerase (TIM) from Methanocaldococcus jannaschii | | Descriptor: | Triosephosphate isomerase | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2006-06-01 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of triosephosphate isomerase (TIM) from Methanocaldococcus jannaschii
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4A6J
 
 | | Structural model of ParM filament based on CryoEM map | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Gayathri, P, Fujii, T, Moller-Jensen, J, Van Den Ent, F, Namba, K, Lowe, J. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
4A62
 
 | |
1ZYO
 
 | | Crystal Structure of the Serine Protease Domain of Sesbania Mosaic Virus polyprotein | | Descriptor: | GLYCEROL, serine protease | | Authors: | Gayathri, P, Satheshkumar, P.S, Prasad, K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2005-06-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the serine protease domain of Sesbania mosaic virus polyprotein and mutational analysis of residues forming the S1-binding pocket
Virology, 346, 2006
|
|
2VFG
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase with 3-phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFD
 
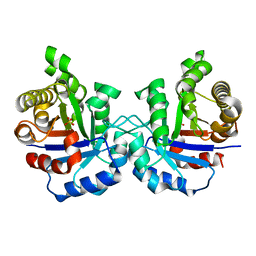 | | Crystal structure of the F96S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFH
 
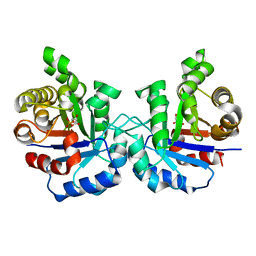 | | Crystal structure of the F96W mutant of Plasmodium falciparum triosephosphate isomerase complexed with 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFF
 
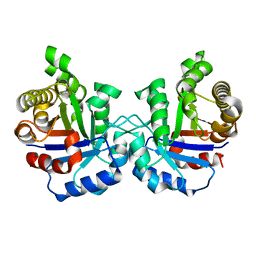 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFI
 
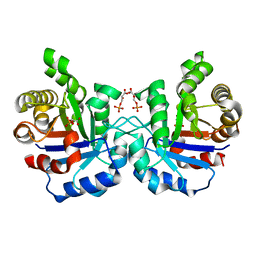 | | Crystal structure of the Plasmodium falciparum triosephosphate isomerase in the loop closed state with 3-phosphoglycerate bound at the active site and interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFA
 
 | | Crystal structure of a chimera of Plasmodium falciparum and human hypoxanthine-guanine phosphoribosyl transferases | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE-XANTHINE PHOSPHORIBOSYLTRANSFERASE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Gayathri, P, Selvi, T.S, Subbayya, I.N.S, Ashok, C.S, Balaram, H, Murthy, M.R.N. | | Deposit date: | 2007-11-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Chimera of Human and Plasmodium Falciparum Hypoxanthine Guanine Phosphoribosyltransferases Provides Insights Into Oligomerization.
Proteins, 73, 2008
|
|
4A61
 
 | | ParM from plasmid R1 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | van den Ent, F, Moller-Jensen, J, Gayathri, P, Lowe, J. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
1P9B
 
 | | Structure of fully ligated Adenylosuccinate synthetase from Plasmodium falciparum | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Eaazhisai, K, Jayalakshmi, R, Gayathri, P, Anand, R.P, Sumathy, K, Balaram, H, Murthy, M.R. | | Deposit date: | 2003-05-10 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fully Ligated Adenylosuccinate Synthetase from Plasmodium falciparum.
J.Mol.Biol., 335, 2004
|
|
6IZW
 
 | | Myxococcus xanthus MglA bound to GTP-gamma-S and MglB | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Gliding motility protein MglB, MAGNESIUM ION, ... | | Authors: | Baranwal, J, Gayathri, P. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of a prokaryotic small Ras-like GTPase contributes to cell polarity oscillations in bacterial motility.
Plos Biol., 17, 2019
|
|
7BVY
 
 | |
7BVZ
 
 | | Crystal structure of MreB5 of Spiroplasma citri bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell shape determining protein MreB, MAGNESIUM ION, ... | | Authors: | Pande, V, Bagde, S.R, Gayathri, P. | | Deposit date: | 2020-04-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MreB5 Is a Determinant of Rod-to-Helical Transition in the Cell-Wall-less Bacterium Spiroplasma.
Curr.Biol., 30, 2020
|
|
5YMX
 
 | | Myxococcus xanthus MglA in GDP bound conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Baranwal, J, Gayathri, P. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Allosteric regulation of a prokaryotic small Ras-like GTPase contributes to cell polarity oscillations in bacterial motility.
Plos Biol., 17, 2019
|
|
5Z66
 
 | | Structure of periplasmic trehalase from Diamondback moth gut bacteria complexed with validoxylamine | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, GLYCEROL, Periplasmic trehalase, ... | | Authors: | Harne, S.R, Adhav, A.S, Joshi, R.S, Gayathri, P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic insights into enzymatic catalysis by trehalase from the insect gut endosymbiont Enterobacter cloacae.
Febs J., 286, 2019
|
|
5Z6H
 
 | | Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form | | Descriptor: | GLYCEROL, Periplasmic trehalase, SULFATE ION | | Authors: | Harne, S.R, Adhav, A.S, Joshi, R.S, Gayathri, P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into enzymatic catalysis by trehalase from the insect gut endosymbiont Enterobacter cloacae.
Febs J., 286, 2019
|
|
8GRW
 
 | | Spiroplasma melliferum FtsZ F224M bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.40005422 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
7YSZ
 
 | | Spiroplasma melliferum FtsZ bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3000412 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
7YOP
 
 | | Spiroplasma melliferum FtsZ bound to GMPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.20003128 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
