2HPT
 
 | | Crystal Structure of E. coli PepN (Aminopeptidase N)in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, GLYCEROL, ... | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPO
 
 | | Structure of Aminopeptidase N from E. coli Suggests a Compartmentalized, Gated Active Site | | Descriptor: | Aminopeptidase N, GLYCEROL, ZINC ION | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1P5C
 
 | |
1OYU
 
 | |
1P56
 
 | |
3EOJ
 
 | | Fmo protein from Prosthecochloris Aestuarii 2K AT 1.3A Resolution | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tronrud, D.E, Wen, J, Gay, L, Blankenship, R.E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
3B2X
 
 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | Descriptor: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | Authors: | Addlagatta, A, Gay, L, Matthews, B.W. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
5UQV
 
 | |
5UQX
 
 | |
2F47
 
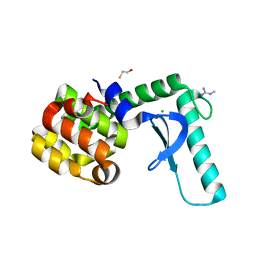 | | Xray crystal structure of T4 lysozyme mutant L20/R63A liganded to methylguanidinium | | Descriptor: | 1-METHYLGUANIDINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-22 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
2F2Q
 
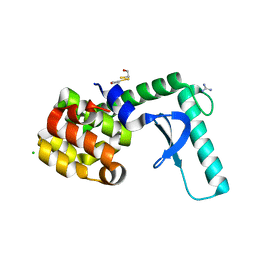 | | High resolution crystal structure of T4 lysozyme mutant L20R63/A liganded to guanidinium ion | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
2F32
 
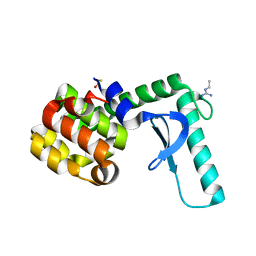 | | Xray crystal structure of lysozyme mutant L20/R63A liganded to ethylguanidinium | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, N-ETHYLGUANIDINE | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
3B3B
 
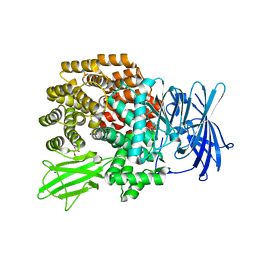 | |
3B37
 
 | |
3B34
 
 | |
3B2P
 
 | | Crystal structure of E. coli Aminopeptidase N in complex with arginine | | Descriptor: | ARGININE, Aminopeptidase N, GLYCEROL, ... | | Authors: | Anthony, A, Leslie, G, Matthews, B.W. | | Deposit date: | 2007-10-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
3JR6
 
 | |
2B7X
 
 | |
1T8A
 
 | | USE OF SEQUENCE DUPLICATION TO ENGINEER A LIGAND-TRIGGERED LONG-DISTANCE MOLECULAR SWITCH IN T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Baase, W.A, Matthews, B.W. | | Deposit date: | 2004-05-11 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T97
 
 | |
5WHC
 
 | | USP7 in complex with Cpd2 (4-(3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl)phenol) | | Descriptor: | 4-[3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl]phenol, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-07-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques.
J. Med. Chem., 60, 2017
|
|
7MSA
 
 | | GDC-9545 in complex with estrogen receptor alpha | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, 3-[(1R,3R)-1-(2,6-difluoro-4-{[1-(3-fluoropropyl)azetidin-3-yl]amino}phenyl)-3-methyl-1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl]-2,2-difluoropropan-1-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zbieg, J.R, Wang, X, Ortwine, D.F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | GDC-9545 (Giredestrant): A Potent and Orally Bioavailable Selective Estrogen Receptor Antagonist and Degrader with an Exceptional Preclinical Profile for ER+ Breast Cancer.
J.Med.Chem., 64, 2021
|
|
