2MU0
 
 | |
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2O5F
 
 | | Crystal Structure of DR0079 from Deinococcus radiodurans at 1.9 Angstrom Resolution | | Descriptor: | Putative Nudix hydrolase DR_0079 | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Characterization of DR_0079 from Deinococcus radiodurans, a Novel Nudix Hydrolase with a Preference for Cytosine (Deoxy)ribonucleoside 5'-Di- and Triphosphates.
Biochemistry, 47, 2008
|
|
3CSX
 
 | |
2OJ1
 
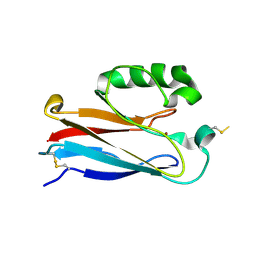 | | Disulfide-linked dimer of azurin N42C/M64E double mutant | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | de Jongh, T.E, Hoffmann, M, Einsle, O, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2007-01-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inter- and intramolecular electron transfer in modified azurin dimers
Eur.J.Inorg.Chem., 2007, 2007
|
|
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
2PNB
 
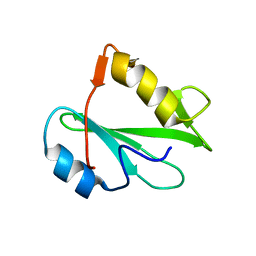 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
2PNI
 
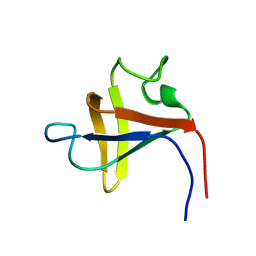 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
2PZY
 
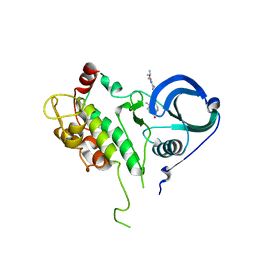 | | Structure of MK2 Complexed with Compound 76 | | Descriptor: | (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE, MAP kinase-activated protein kinase 2, STAUROSPORINE | | Authors: | White, A, Wu, J.P, Wang, J, Abeywardane, A, Andersen, D, Emmanuel, M, Gautschi, E, Goldberg, D.R, Kashem, M.A, Lukas, S, Mao, W, Martin, L, Morwick, T, Moss, N, Pargellis, C, Patel, U.R, Patnaude, L, Peet, G.W, Skow, D, Snow, R.J, Ward, Y, Werneburg, B. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The discovery of carboline analogs as potent MAPKAP-K2 inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1R75
 
 | |
1SKN
 
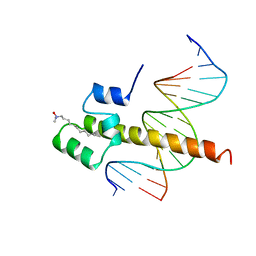 | | THE BINDING DOMAIN OF SKN-1 IN COMPLEX WITH DNA: A NEW DNA-BINDING MOTIF | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*GP*AP*TP*GP*AP*CP*AP*TP*TP*GP*T)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*TP*CP*CP*C)-3'), DNA-BINDING DOMAIN OF SKN-1, ... | | Authors: | Rupert, P.B, Daughdrill, G.W, Bowerman, B, Matthews, B.W. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new DNA-binding motif in the Skn-1 binding domain-DNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1SSV
 
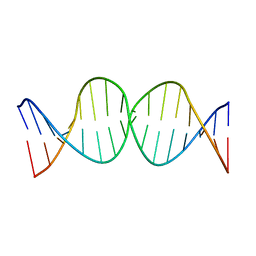 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1SS7
 
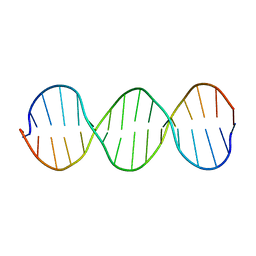 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1U49
 
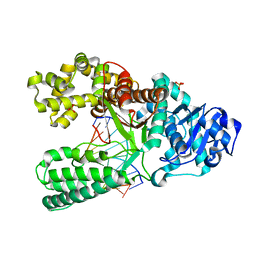 | | Adenine-8oxoguanine mismatch at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U48
 
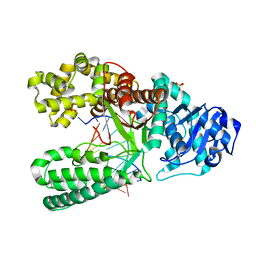 | | Extension of a cytosine-8-oxoguanine base pair | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U45
 
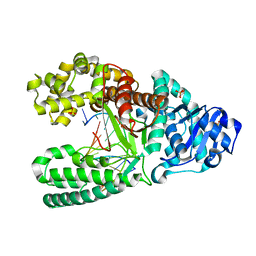 | | 8oxoguanine at the pre-insertion site of the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1TTL
 
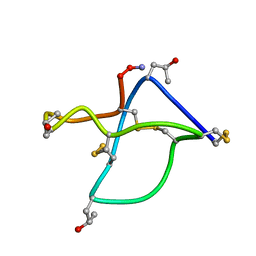 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1U4B
 
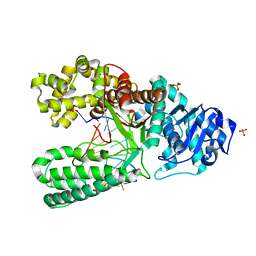 | | Extension of an adenine-8oxoguanine mismatch | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1UA0
 
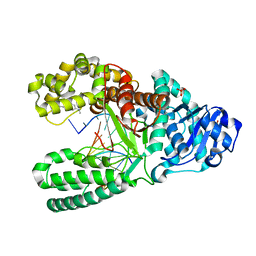 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1TR6
 
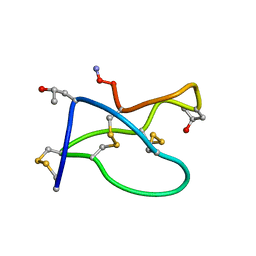 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1UA1
 
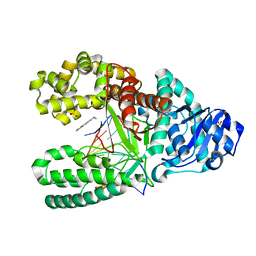 | | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1U47
 
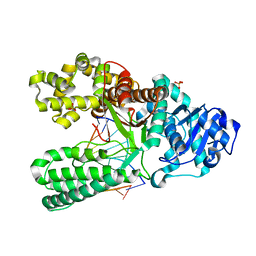 | | cytosine-8-Oxoguanine base pair at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1TPT
 
 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
1VJ6
 
 | |
