6TTD
 
 | | Crystal structure of McoA multicopper oxidase 2F4 variant from the hyperthermophile Aquifex aeolicus | | Descriptor: | COPPER (II) ION, GLYCEROL, Multicopper oxidase, ... | | Authors: | Borges, P.T, Brissos, V, Cordeiro, T.N, Martins, L.O, Frazao, C. | | Deposit date: | 2019-12-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
Acs Catalysis, 2022
|
|
7QYZ
 
 | | Crystal structure of a DyP-type peroxidase 6E10 variant from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Martins, L.O, Frazao, C. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7QZA
 
 | | Crystal structure of a DyP-type peroxidase 29E4 variant from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7QYQ
 
 | | Crystal structure of a DyP-type peroxidase from Pseudomonas putida | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Silva, D, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Unveiling molecular details behind improved activity at neutral to alkaline pH of an engineered DyP-type peroxidase.
Comput Struct Biotechnol J, 20, 2022
|
|
7R0F
 
 | | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2P
 
 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2O
 
 | | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R1J
 
 | | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R2S
 
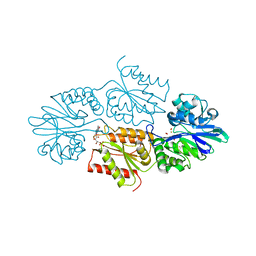 | | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas
To Be Published
|
|
7R1H
 
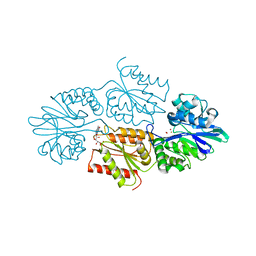 | | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R2R
 
 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
1CTJ
 
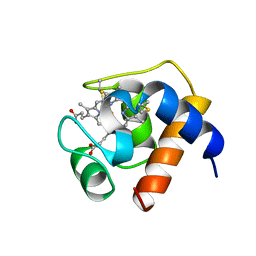 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sheldrick, G.M. | | Deposit date: | 1995-08-08 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ab initio determination of the crystal structure of cytochrome c6 and comparison with plastocyanin.
Structure, 3, 1995
|
|
1BBS
 
 | |
1SMR
 
 | |
3CYR
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774P | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | Deposit date: | 1997-07-24 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the Three-Dimensional Structures of Cytochrome C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstroms Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstroms Resolution
Inorg.Chim.Acta., 273, 1998
|
|
1W9M
 
 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2CTH
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | Deposit date: | 1997-06-18 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Refinement of the Three-Dimensional Structures of Cytochromes C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstrom Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstrom Resolution
Inorg.Chim.Acta., 273, 1998
|
|
1MDV
 
 | | KEY ROLE OF PHENYLALANINE 20 IN CYTOCHROME C3: STRUCTURE, STABILITY AND FUNCTION STUDIES | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dolla, A, Arnoux, P, Protasevich, I, Lobachov, V, Brugna, M, Guidici-Orticoni, M.T, Haser, R, Czjzek, M, Makarov, A, Brushi, M. | | Deposit date: | 1998-09-08 | | Release date: | 1999-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key role of phenylalanine 20 in cytochrome c3: structure, stability, and function studies.
Biochemistry, 38, 1999
|
|
1UPD
 
 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1UP9
 
 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1CZI
 
 | | CHYMOSIN COMPLEX WITH THE INHIBITOR CP-113972 | | Descriptor: | CHYMOSIN, CP-113972 (NORSTATINE-S-METHYL CYSTEINE-IODO-PHENYLALANINE-PROLINE) | | Authors: | Groves, M.R, Dhanaraj, V, Pitts, J.E, Badasso, M, Hoover, D, Nugent, P, Blundell, T.L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure.
Protein Eng., 11, 1998
|
|
1CED
 
 | | THE STRUCTURE OF CYTOCHROME C6 FROM MONORAPHIDIUM BRAUNII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Quacquarini, G, Walter, O, Diaz, A, Hervas, M, De La Rosa, M.A. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of cytochrome c6 from the green alga Monoraphidium braunii
J.Biol.Inorg.Chem., 1, 1996
|
|
1GMB
 
 | | Reduced structure of CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome
J.Biol.Chem., 276, 2001
|
|
1GM4
 
 | | OXIDISED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-10 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome.
J.Biol.Chem., 276, 2001
|
|
