6CT4
 
 | | TFE-induced NMR structure of an antimicrobial peptide (EcDBS1R5) derived from a mercury transporter protein (MerP - Escherichia coli) | | Descriptor: | EcDBS1R5 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Computationally Designed Peptide Derived from Escherichia coli as a Potential Drug Template for Antibacterial and Antibiofilm Therapies.
ACS Infect Dis, 4, 2018
|
|
6CT1
 
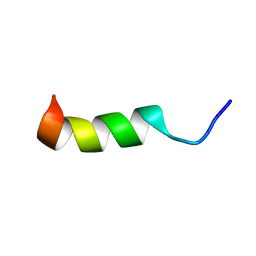 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R7) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R7 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
6CSZ
 
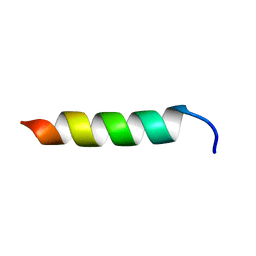 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R3) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R3 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
6CSK
 
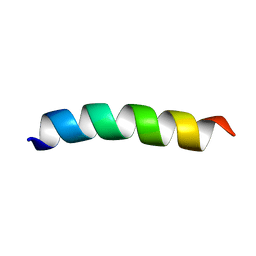 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R2) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R2 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-2 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2N0V
 
 | | Backbone 1H, Chemical Shift Assignments for Cn-APM1 | | Descriptor: | Antimicrobial peptide 1 | | Authors: | Santana, M.J, Oliveira, A.L, Queiroz Jr, L.K, Mandal, S.M, Matos, C.O, Dias, R.O, Franco, O.L, Liao, L.M. | | Deposit date: | 2015-03-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water.
Febs Lett., 589, 2015
|
|
8EP5
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1 | | Descriptor: | Mastoparan-R1 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1.
To Be Published
|
|
8ERU
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R4 | | Descriptor: | Mastoparan-R4 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computacionally designed mastoparan-like peptide, mastoparan-R4
To Be Published
|
|
5V1E
 
 | |
7UNX
 
 | |
6CTG
 
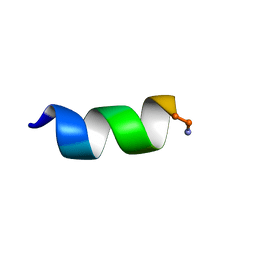 | |
8T3N
 
 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
6C41
 
 | |
6D2H
 
 | |
2MP9
 
 | | Solution structure of an potent antifungal peptide Cm-p5 derived from C. muricatus | | Descriptor: | Antifungal peptide | | Authors: | Sun, Z.J, Heffron, G, Mcbeth, C, Wagner, G, Otero-Gonzales, A.J, Starnbach, M.N. | | Deposit date: | 2014-05-14 | | Release date: | 2015-08-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cm-p5: an antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae).
Faseb J., 29, 2015
|
|
2N9R
 
 | | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics | | Descriptor: | Antimicrobial peptide PaDBS1R1 | | Authors: | Irazazabal, L.S.F, Porto, W.F, Alves, E.S.F, Matos, C.O, Hancock, R.E.W, Haney, E, Ribeiro, S.M, Liao, L.M, Ladram, A.S.F, Franco, O.L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-07 | | Method: | SOLUTION NMR | | Cite: | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics
To be Published
|
|
6E7K
 
 | |
