2X28
 
 | | cadmium bound structure of SporoSAG | | Descriptor: | CADMIUM ION, SPOROZOITE-SPECIFIC SAG PROTEIN | | Authors: | Crawford, J, Lamb, E, Wasmuth, J, Grujic, O, Grigg, M.E, Boulanger, M.J. | | Deposit date: | 2010-01-11 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Sporosag: A Sag2 Related Surface Antigen from Toxoplasma Gondii.
J.Biol.Chem., 285, 2010
|
|
2WNK
 
 | | Structure of SporoSAG from Toxoplasma gondii | | Descriptor: | SPOROZOITE-SPECIFIC SAG PROTEIN | | Authors: | Crawford, J, Lamb, E, Grujic, O, Grigg, M.E, Boulanger, M.J. | | Deposit date: | 2009-07-09 | | Release date: | 2010-02-02 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of Sporosag: A Sag2 Related Surface Antigen from Toxoplasma Gondii.
J.Biol.Chem., 285, 2010
|
|
2X2Z
 
 | | Crystal structure AMA1 from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN 1, PUTATIVE, ... | | Authors: | Crawford, J, Tonkin, M.L, Grujic, O, Boulanger, M.J. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Apical Membrane Antigen 1 (Ama1) from Toxoplasma Gondii.
J.Biol.Chem., 285, 2010
|
|
1KQQ
 
 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
8E1O
 
 | | Crystal structure of hTEAD2 bound to a methoxypyridine lipid pocket binder | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methoxy-N-({3-[2-(methylamino)-2-oxoethyl]phenyl}methyl)-4-{(E)-2-[trans-4-(trifluoromethyl)cyclohexyl]ethenyl}pyridine-2-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Dey, A, Zbieg, J, Crawford, J. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeting the Hippo pathway in cancers via ubiquitination dependent TEAD degradation
Biorxiv, 2024
|
|
2Y8R
 
 | | Crystal structure of apo AMA1 mutant (Tyr230Ala) from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
4XE4
 
 | | Coagulation Factor XII protease domain crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL | | Authors: | Pathak, M, Wilmann, P, Awford, J, Li, C, Fisher, P.M, Dreveny, I, Dekker, L.V, Emsley, J. | | Deposit date: | 2014-12-22 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coagulation factor XII protease domain crystal structure.
J.Thromb.Haemost., 13, 2015
|
|
4XDE
 
 | | Coagulation Factor XII protease domain crystal structure | | Descriptor: | CITRATE ANION, Coagulation factor XII, ISOPROPYL ALCOHOL | | Authors: | Pathak, M, Wilmann, P, Awford, J, Li, C, Fisher, P.M, Dreveny, I, Dekker, L.V, Emsley, J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Coagulation factor XII protease domain crystal structure.
J.Thromb.Haemost., 13, 2015
|
|
2Y8T
 
 | | Co-structure of AMA1 with a surface exposed region of RON2 from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE, ... | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
2Y8S
 
 | | Co-structure of an AMA1 mutant (Y230A) with a surface exposed region of RON2 from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE, ... | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
5FUF
 
 | | Crystal structure of the Mep2 mutant S453D from Candida albicans | | Descriptor: | AMMONIUM TRANSPORTER, DECYL-BETA-D-MALTOPYRANOSIDE | | Authors: | van den Berg, B, Chembath, A, Jefferies, D, Basle, A, Khalid, S, Rutherford, J. | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Structural Basis for Mep2 Ammonium Transceptor Activation by Phosphorylation.
Nat.Commun., 7, 2016
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
5AID
 
 | |
5AH3
 
 | |
4APM
 
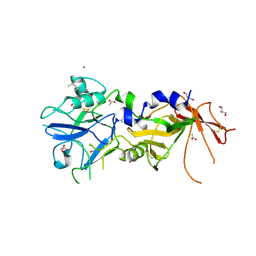 | | Crystal Structure of AMA1 from Babesia divergens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, APICAL MEMBRANE ANTIGEN 1, ... | | Authors: | Tonkin, M.L, Crawford, J, Lebrun, M.L, Boulanger, M.J. | | Deposit date: | 2012-04-04 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Babesia Divergens and Neospora Caninum Apical Membrane Antigen 1 (Ama1) Structures Reveal Selectivity and Plasticity in Apicomplexan Parasite Host Cell Invasion.
Protein Sci., 22, 2013
|
|
4APL
 
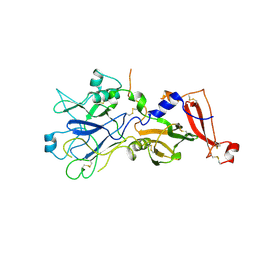 | |
2XGG
 
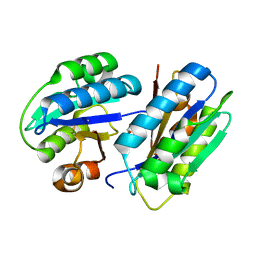 | | Structure of Toxoplasma gondii Micronemal Protein 2 A_I Domain | | Descriptor: | GLYCEROL, MICRONEME PROTEIN 2 | | Authors: | Tonkin, M.L, Grujic, O, Pearce, M, Crawford, J, Boulanger, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Micronemal Protein 2 (Mic2) A/I Domain from Toxoplasma Gondii.
Protein Sci., 19, 2010
|
|
2JKS
 
 | | Crystal structure of the the bradyzoite specific antigen BSR4 from toxoplasma gondii. | | Descriptor: | BRADYZOITE SURFACE ANTIGEN BSR4, ZINC ION | | Authors: | Boulanger, M.J, Grigg, M.E, Bruic, E, Grujic, O. | | Deposit date: | 2008-08-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of the Bradyzoite Surface Antigen (Bsr4) from Toxoplasma Gondii, a Unique Addition to the Surface Antigen Glycoprotein 1-Related Superfamily.
J.Biol.Chem., 284, 2009
|
|
8DOV
 
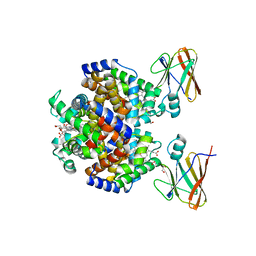 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2M3C
 
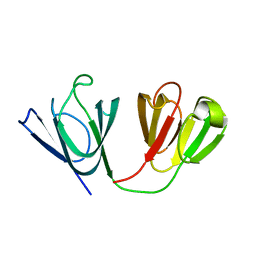 | | Solution Structure of gammaM7-Crystallin | | Descriptor: | Crystallin, gamma M7 | | Authors: | Mahler, B, Wu, Z. | | Deposit date: | 2013-01-16 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Fish Eye Lens Protein, gamma M7-Crystallin.
Biochemistry, 52, 2013
|
|
3V5L
 
 | |
4FPO
 
 | |
4FP2
 
 | |
4FPL
 
 | |
