6KG6
 
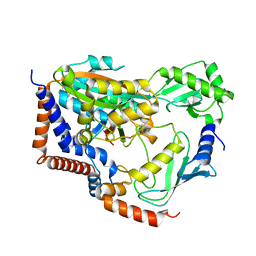 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
8I24
 
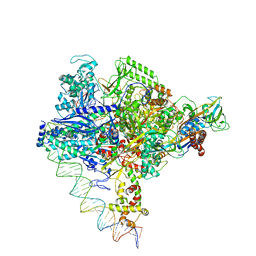 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
7WE6
 
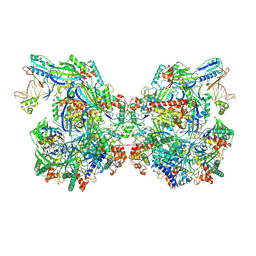 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7X0R
 
 | |
7X0I
 
 | |
7X0N
 
 | |
7X0M
 
 | |
7X0L
 
 | |
7X0K
 
 | |
7X0P
 
 | |
7X0J
 
 | |
7X0O
 
 | |
7X0Q
 
 | |
7X0H
 
 | |
7X0G
 
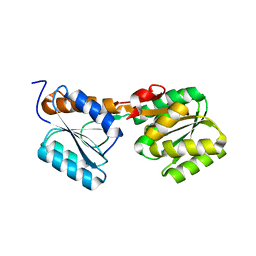 | |
7YHI
 
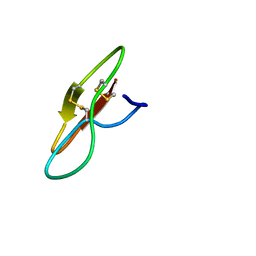 | |
7YHG
 
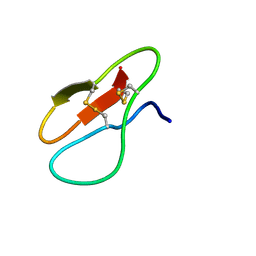 | |
7YHH
 
 | |
7YHF
 
 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECW
 
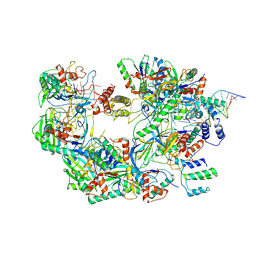 | | The Csy-AcrIF14-dsDNA complex | | Descriptor: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7C6S
 
 | |
