2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
2GF0
 
 | | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state | | Descriptor: | GTP-binding protein Di-Ras1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Turnbull, A.P, Papagrigoriou, E, Yang, X, Schoch, G, Elkins, J, Gileadi, O, Salah, E, Bray, J, Wen-Hwa, L, Fedorov, O, Niesen, F.E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state
To be Published
|
|
2G45
 
 | | Co-crystal structure of znf ubp domain from the deubiquitinating enzyme isopeptidase T (isot) in complex with ubiquitin | | Descriptor: | CHLORIDE ION, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2G43
 
 | | Structure of the ZNF UBP domain from deubiquitinating enzyme isopeptidase T (IsoT) | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ZINC ION | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2GMJ
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase | | Descriptor: | Electron transfer flavoprotein-ubiquinone oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GMH
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase in Complexed with Ubiquinone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, Electron transfer flavoprotein-ubiquinone oxidoreductase, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HR5
 
 | | PF1283- Rubrerythrin from Pyrococcus furiosus iron bound form | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Dillard, B.D, Ruble, J.R, Chen, L, Liu, Z.J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of iron bound Rubrerythrin from Pyrococcus Furiosus
To be Published
|
|
2I47
 
 | | Crystal structure of catalytic domain of TACE with inhibitor | | Descriptor: | 4-({[4-(BUT-2-YN-1-YLOXY)PHENYL]SULFONYL}METHYL)-1-[(3,5-DIMETHYLISOXAZOL-4-YL)SULFONYL]-N-HYDROXYPIPERIDINE-4-CARBOXAMIDE, ADAM 17, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Xu, W, Condon, J.S, Lovering, F.E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent and selective TACE inhibitors via the S1 pocket.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2K1P
 
 | |
2LHO
 
 | | Solution Structure of a DNA duplex Containing an Unnatural, Hydrophobic Base Pair | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*TP*CP*(LHO)P*TP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*AP*(MM7)P*GP*AP*AP*AP*CP*G)-3') | | Authors: | Malyshev, D.A, Pfaff, D.A, Ippoliti, S.L, Hwang, G.T, Dwyer, T.J, Romesberg, F.E. | | Deposit date: | 2011-08-12 | | Release date: | 2012-07-04 | | Method: | SOLUTION NMR | | Cite: | Solution structure, mechanism of replication, and optimization of an unnatural base pair.
Chemistry, 16, 2010
|
|
2LI8
 
 | |
1SGW
 
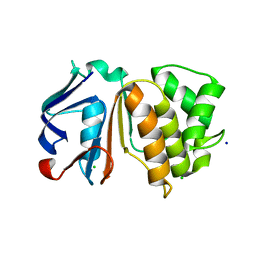 | | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative ABC transporter | | Authors: | Liu, Z.J, Tempel, W, Shah, A, Chen, L, Lee, D, Kelley, L.-L.C, Dillard, B.D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001
To be Published
|
|
1TWL
 
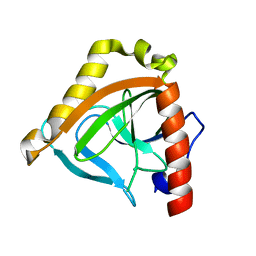 | | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Zhou, W, Tempel, W, Liu, Z.-J, Chen, L, Clancy Kelley, L.-L, Dillard, B.D, Hopkins, R.C, Arendall III, W.B, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-07-01 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001
To be published
|
|
1UM4
 
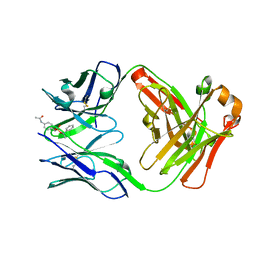 | | Catalytic Antibody 21H3 with hapten | | Descriptor: | (1R)-1-PHENYLETHYL 4-(ACETYLAMINO)BENZYLPHOSPHONATE, Antibody 21H3 H chain, Antibody 21H3 L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
1UM6
 
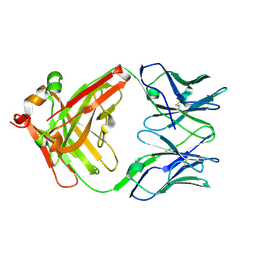 | | catalytic antibody 21h3 | | Descriptor: | antibody 21h3, H chain, L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
1UM5
 
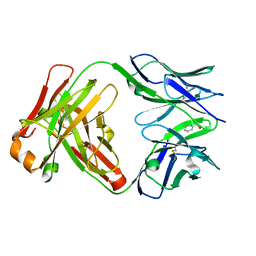 | | Catalytic Antibody 21H3 with alcohol substrate | | Descriptor: | 1-PHENYLETHANOL, Antibody 21H3 H chain, Antibody 21H3 L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
1VCX
 
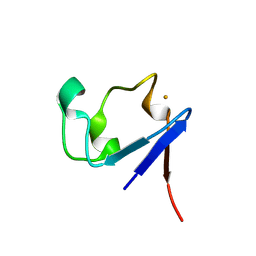 | | Neutron Crystal Structure of the Wild Type Rubredoxin from Pyrococcus Furiosus at 1.5A Resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Kurihara, K, Tanaka, I, Chatake, T, Adams, M.W.W, Jenney Jr, F.E, Moiseeva, N, Bau, R, Niimura, N. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VKC
 
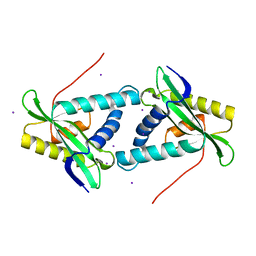 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
1VJK
 
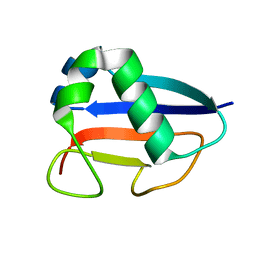 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
1XHC
 
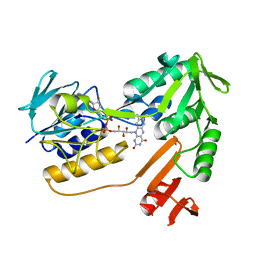 | | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase /nitrite reductase | | Authors: | Horanyi, P, Tempel, W, Weinberg, M.V, Liu, Z.-J, Shah, A, Chen, L, Lee, D, Sugar, F.J, Brereton, P.S, Izumi, M, Poole II, F.L, Shah, C, Jenney Jr, F.E, Arendall III, W.B, Rose, J.P, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-17 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001
To be published
|
|
1XG7
 
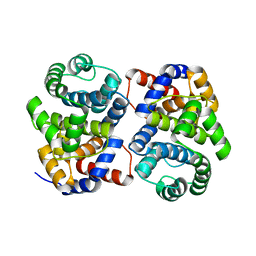 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1XI9
 
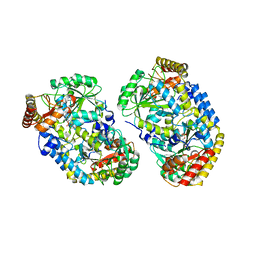 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
1XI3
 
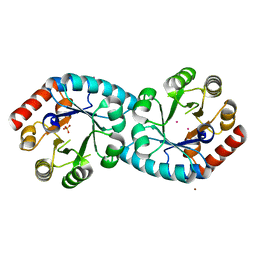 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
1XX7
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001 | | Descriptor: | NICKEL (II) ION, UNKNOWN ATOM OR ION, oxetanocin-like protein | | Authors: | Chen, L, Tempel, W, Habel, J, Zhou, W, Nguyen, D, Chang, S.-H, Lee, D, Kelley, L.-L.C, Dillard, B.D, Liu, Z.-J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001
To be published
|
|
1Y0J
 
 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
