4TX9
 
 | | Crystal structure of HisAp from Streptomyces sviceus with degraded ProFAR | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Phosphoribosyl isomerase A, SULFATE ION | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4GB5
 
 | | Crystal structure of Kfla4162 protein from Kribbella flavida | | Descriptor: | PHOSPHATE ION, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Michalska, K, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Kfla4162 protein from Kribbella flavida (CASP Target)
To be Published
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
4W9T
 
 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4WD0
 
 | | Crystal structure of HisAp form Arthrobacter aurescens | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of HisAp form Arthrobacter aurescens
To Be Published
|
|
4WIW
 
 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4WUI
 
 | | Crystal structure of TrpF from Jonesia denitrificans | | Descriptor: | CITRIC ACID, N-(5'-phosphoribosyl)anthranilate isomerase | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5E7Q
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6OK3
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ONW
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORC
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORK
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6OSS
 
 | | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHITE ION, SULFATE ION | | Authors: | Kim, Y, Stogios, P, Skarina, T, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis
To Be Published
|
|
4G6Q
 
 | | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Tan, K, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836, CASP Target
To be Published
|
|
4GAK
 
 | | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale | | Descriptor: | Acyl-ACP thioesterase, CHLORIDE ION, GLYCEROL | | Authors: | Chang, C, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale
To be Published
|
|
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
5E2E
 
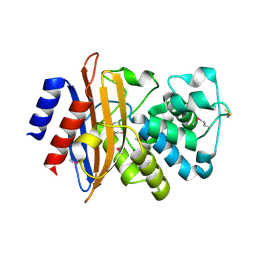 | | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica
To Be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
5DN1
 
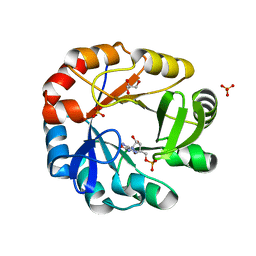 | | Crystal structure of Phosphoribosyl isomerase A from Streptomyces coelicolor | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, GLYCEROL, Phosphoribosyl isomerase A, ... | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-09 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5E2F
 
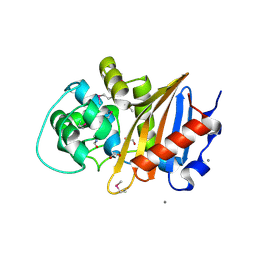 | | Crystal Structure of Beta-lactamase class D from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase YbxI, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Beta-lactamase class D from Bacillus subtilis
To Be Published
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
6X9Y
 
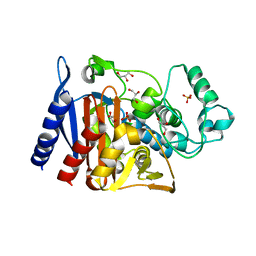 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6XG1
 
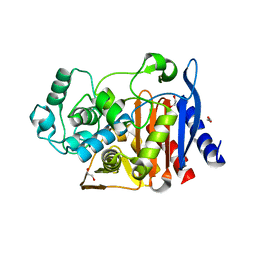 | | Class C beta-lactamase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
6XG3
 
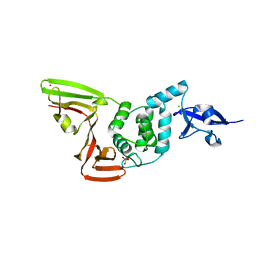 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | Descriptor: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
