4C7N
 
 | | Crystal Structure of the synthetic peptide iM10 in complex with the coiled-coil region of MITF | | Descriptor: | MERCURY (II) ION, MICROPHTHALMIA ASSOCIATED TRANSCRIPTION FACTOR, SYNTHETIC ALPHA-HELIX, ... | | Authors: | Wohlwend, D, Gerhardt, S, Kuekenshoener, T, Einsle, O. | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving Coiled Coil Stability While Maintaining Specificity by a Bacterial Hitchhiker Selection System.
J.Struct.Biol., 186, 2014
|
|
4FC4
 
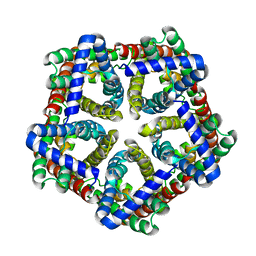 | | FNT family ion channel | | Descriptor: | Nitrite transporter NirC, octyl beta-D-glucopyranoside | | Authors: | Lue, W, Schwarzer, N, Du, J, Gerbig-Smentek, E, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2012-05-24 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7NDM
 
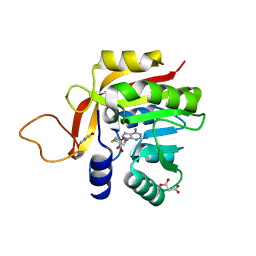 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound substrate 4-hydroxyisoquinolin-1(2H)-one | | Descriptor: | 4-oxidanyl-2~{H}-isoquinolin-1-one, Heterocyclic toxin methyltransferase (Rv0560c), MALONATE ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7NOY
 
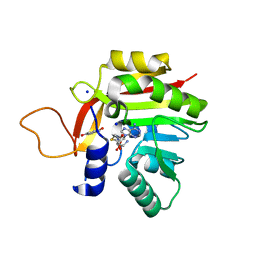 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis in complex with substrate 1-hydroxyquinolin-4(1H)-one | | Descriptor: | 1-oxidanylquinolin-4-one, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7NMK
 
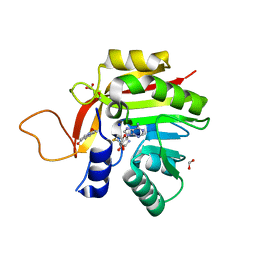 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound methylation product 1-methoxyquinolin-4(1H)-one | | Descriptor: | 1-methoxy-4-oxoquinoline, 2-heptyl-1-hydroxyquinolin-4(1H)-one methyltransferase, FORMIC ACID, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
4WNA
 
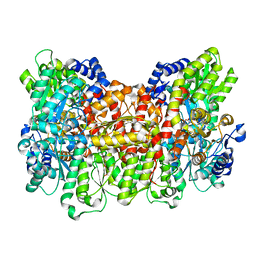 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
6HBD
 
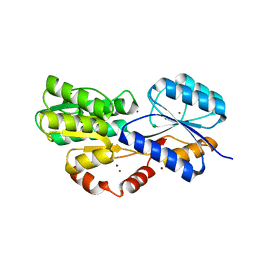 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Galactofuranose | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION, beta-D-galactofuranose | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HBM
 
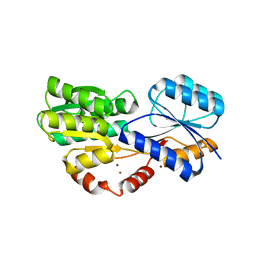 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with alpha-L-arabinofuranose | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION, alpha-L-arabinofuranose | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HB0
 
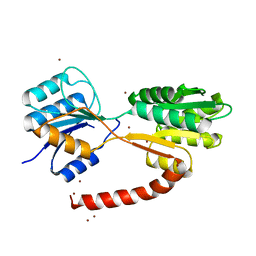 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HYH
 
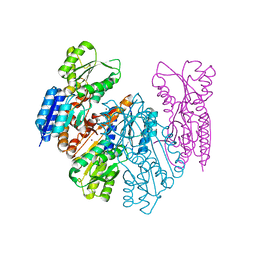 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | Authors: | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | Deposit date: | 2015-01-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
1JVL
 
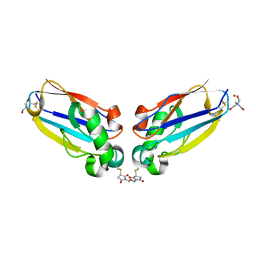 | | Azurin dimer, covalently crosslinked through bis-maleimidomethylether | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
1JVO
 
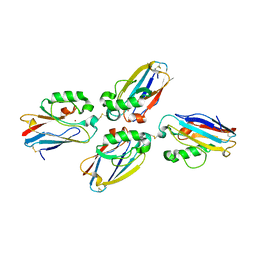 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
7ZNQ
 
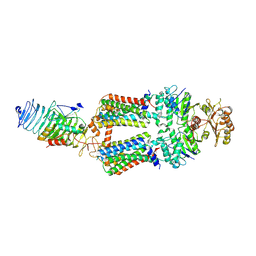 | | ABC transporter complex NosDFYL in GDN | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Zhang, L, Mueller, C, Zipfel, S, Chami, M, Einsle, O. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
8AHX
 
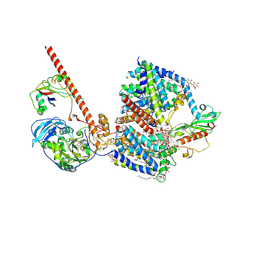 | |
8BOQ
 
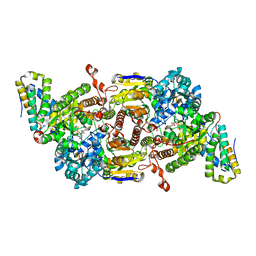 | | A. vinelandii Fe-nitrogenase FeFe protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE(8)-S(7) CLUSTER, Fe-only nitrogenase, ... | | Authors: | Trncik, C, Detemple, F, Einsle, O. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Iron-only Fe-nitrogenase underscores common catalytic principles in biological nitrogen fixation
Nat Catal, 2023
|
|
8BCX
 
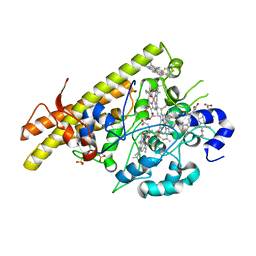 | |
8CE1
 
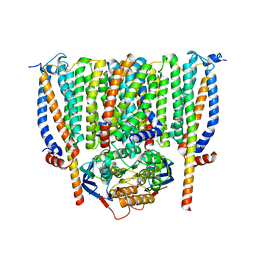 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE5
 
 | | Cytochrome c maturation complex CcmABCD, E154Q, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CEA
 
 | | Cytochrome c maturation complex CcmABCD, E154Q | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE8
 
 | | Cytochrome c maturation complex CcmABCDE | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Cytochrome c-type biogenesis protein CcmE, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
3DP5
 
 | | Crystal structure of Geobacter sulfurreducens OmcF with N-terminal Strep-tag II | | Descriptor: | Cytochrome c family protein, HEME C, SULFATE ION | | Authors: | Lukat, P, Hoffmann, M, Einsle, O. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-02 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal packing of the c(6)-type cytochrome OmcF from Geobacter sulfurreducens is mediated by an N-terminal Strep-tag II
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
3HQ7
 
 | | CcpA from G. sulfurreducens, G94K/K97Q/R100I variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ6
 
 | | Cytochrome c peroxidase from G. sulfurreducens, wild type | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ8
 
 | | CcpA from G. sulfurreducens S134P/V135K variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, IMIDAZOLE, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
