3HQ6
 
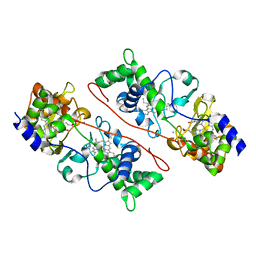 | | Cytochrome c peroxidase from G. sulfurreducens, wild type | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
5DY4
 
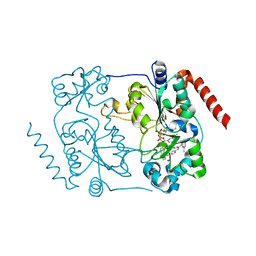 | | Crystal structure of human Sirt2 in complex with a brominated 2nd generation SirReal inhibitor and NAD+ | | Descriptor: | N-{5-[(7-bromonaphthalen-1-yl)methyl]-1,3-thiazol-2-yl}-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aminothiazoles as Potent and Selective Sirt2 Inhibitors: A Structure-Activity Relationship Study.
J.Med.Chem., 59, 2016
|
|
4L4Q
 
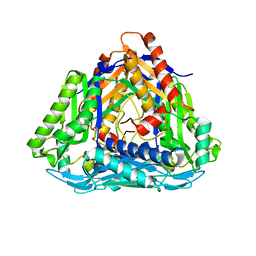 | | Methionine Adenosyltransferase | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Schlesier, J, Siegrist, J, Gerhardt, S, Andexer, J.N, Einsle, O. | | Deposit date: | 2013-06-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis.
Bmc Struct.Biol., 13, 2013
|
|
4LYW
 
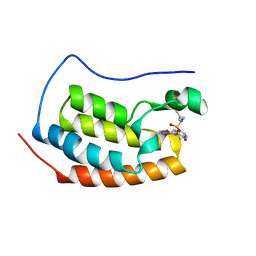 | | Crystal Structure of BRD4(1) bound to inhibitor XD14 | | Descriptor: | 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LZR
 
 | | Crystal Structure of BRD4(1) bound to Colchicine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O, Huegle, M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LZS
 
 | | Crystal Structure of BRD4(1) bound to inhibitor XD46 | | Descriptor: | 4-acetyl-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Einsle, O, Gerhardt, S. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7QBA
 
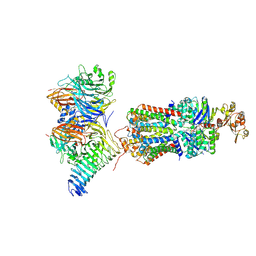 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
4LYS
 
 | | Crystal Structure of BRD4(1) bound to Colchiceine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-10-hydroxy-1,2,3-trimethoxy-9-oxo-5,6,7,9-tetrahydrobenzo[a]heptalen-7-yl]acetamide, SODIUM ION | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7QQA
 
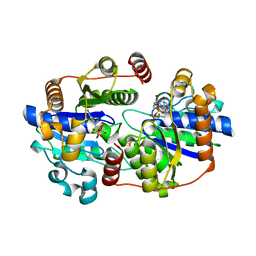 | | MgADP-bound Fe protein of the iron-only nitrogenase from Azotobacter vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Trncik, C, Mueller, T, Franke, P, Einsle, O. | | Deposit date: | 2022-01-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | MgADP-bound Fe protein of the iron-only nitrogenase from Azotobacter vinelandii
Journal of Inorganic Biochemistry, 227, 2022
|
|
4AUP
 
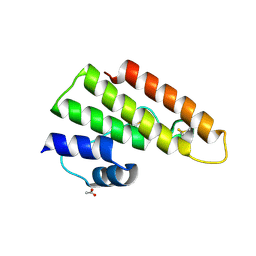 | | Tuber borchii Phospholipase A2 | | Descriptor: | ACETATE ION, PHOSPHOLIPASE A2 GROUP XIII, THIOCYANATE ION | | Authors: | Cavazzini, D, Meschi, F, Corsini, R, Bolchi, A, Rossi, G.-L, Einsle, O, Ottonello, S. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoproteolytic Activation of a Symbiosis-Regulated Truffle Phospholipase A2
J.Biol.Chem., 288, 2013
|
|
4C7N
 
 | | Crystal Structure of the synthetic peptide iM10 in complex with the coiled-coil region of MITF | | Descriptor: | MERCURY (II) ION, MICROPHTHALMIA ASSOCIATED TRANSCRIPTION FACTOR, SYNTHETIC ALPHA-HELIX, ... | | Authors: | Wohlwend, D, Gerhardt, S, Kuekenshoener, T, Einsle, O. | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-02 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving Coiled Coil Stability While Maintaining Specificity by a Bacterial Hitchhiker Selection System.
J.Struct.Biol., 186, 2014
|
|
4B2N
 
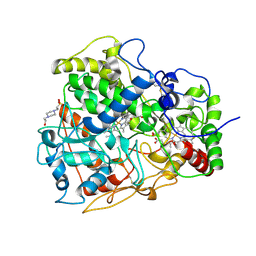 | | Latex Oxygenase RoxA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 70 KDA PROTEIN, HEME C, ... | | Authors: | Seidel, J, Schmitt, G, Hoffmann, M, Jendrossek, D, Einsle, O. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the processive rubber oxygenase RoxA from Xanthomonas sp.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
5AFW
 
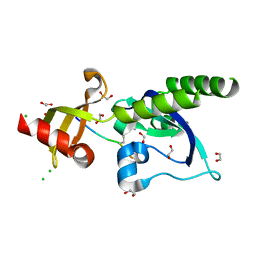 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5AR1
 
 | | Crystal structure of Cdc11 from Saccharomyces cerevisiae | | Descriptor: | CELL DIVISION CONTROL PROTEIN 11 | | Authors: | Brausemann, A, Gerhardt, S, Schott, A.K, Einsle, O, Grosse-Berkenbusch, A, Johnsson, N, Gronemeyer, T. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Cdc11, a Septin Subunit from Saccharomyces Cerevisiae.
J.Struct.Biol., 193, 2016
|
|
5D7O
 
 | | Crystal structure of Sirt2-ADPR at an improved resolution | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5D7N
 
 | | Crystal structure of human Sirt3 at an improved resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2CZS
 
 | |
2B2I
 
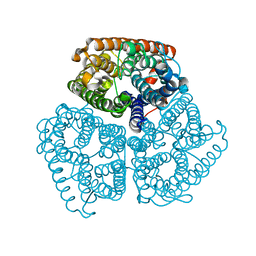 | |
2B2J
 
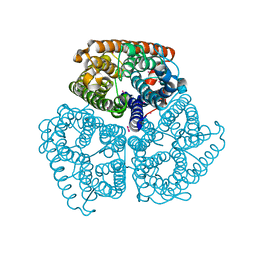 | | Ammonium Transporter Amt-1 from A. fulgidus (Xe) | | Descriptor: | XENON, ammonium transporter | | Authors: | Andrade, S.L.A, Dickmanns, A, Ficner, R, Einsle, O. | | Deposit date: | 2005-09-19 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B2F
 
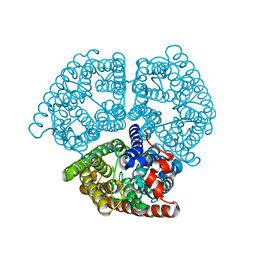 | |
2B2H
 
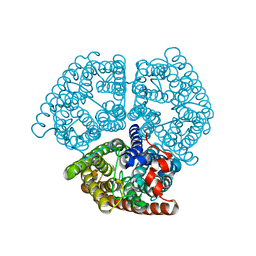 | |
7ADR
 
 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7ADY
 
 | | CO-removed state of the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3B6M
 
 | | WrbA from Escherichia coli, second crystal form | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein wrbA, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Andrade, S.L.A, Patridge, E.V, Ferry, J.G, Einsle, O. | | Deposit date: | 2007-10-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli.
J.Bacteriol., 189, 2007
|
|
3B6K
 
 | | WrbA from Escherichia coli, Benzoquinone complex | | Descriptor: | 1,4-benzoquinone, FLAVIN MONONUCLEOTIDE, Flavoprotein wrbA, ... | | Authors: | Andrade, S.L.A, Patridge, E.V, Ferry, J.G, Einsle, O. | | Deposit date: | 2007-10-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli.
J.Bacteriol., 189, 2007
|
|
