3PBP
 
 | |
3PXM
 
 | | Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II helix | | Descriptor: | Monellin chain B/Monellin chain A chimeric protein | | Authors: | Hobbs, J.R, Templeton, C.M, Pour, S.O, Blanch, E.W, Munger, S.M, Conn, G.L. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reduced Sweetness of a Monellin (MNEI) Mutant Results from Increased Protein Flexibility and Disruption of a Distant Poly-(L-Proline) II Helix.
Chem Senses, 36, 2011
|
|
3QIB
 
 | | Crystal structure of the 2B4 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 beta chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
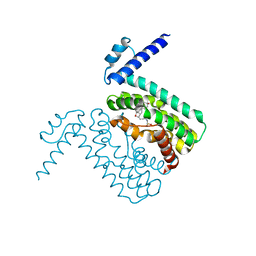 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QJF
 
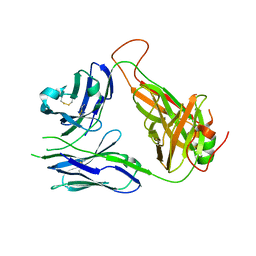 | | Crystal structure of the 2B4 TCR | | Descriptor: | 2B4 alpha chain, 2B4 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QJH
 
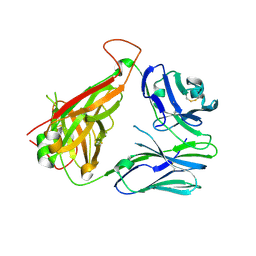 | | The crystal structure of the 5c.c7 TCR | | Descriptor: | 5c.c7 alpha chain, 5c.c7 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QIW
 
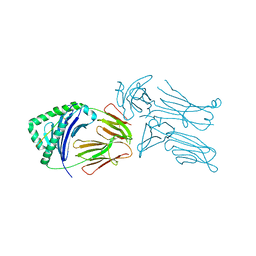 | | Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QIU
 
 | | Crystal structure of the 226 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
4CGL
 
 | | Leishmania major N-myristoyltransferase in complex with an aminoacylpyrrolidine inhibitor | | Descriptor: | (3R)-3-azanyl-4-(4-chlorophenyl)-1-[(3S,4R)-3-(4-chlorophenyl)-4-(hydroxymethyl)pyrrolidin-1-yl]butan-1-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4CGN
 
 | | Leishmania major N-myristoyltransferase in complex with a piperidinylindole inhibitor | | Descriptor: | 2-(4-fluorophenyl)-N-(3-piperidin-4-yl-1H-indol-5-yl)ethanamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
3AUV
 
 | | Predicting Amino Acid Preferences in the Complementarity Determining Regions of an Antibody-Antigen Recognition Interface | | Descriptor: | sc-dsFv derived from the G6-Fab | | Authors: | Yu, C.M, Peng, H.P, Chen, I.C, Lee, Y.C, Chen, J.B, Tsai, K.C, Chen, C.T, Chang, J.Y, Yang, E.W, Hsu, P.C, Jian, J.W, Hsu, H.J, Chang, H.J, Hsu, W.L, Huang, K.F, Ma, A.C, Yang, A.S. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface
Plos One, 7, 2012
|
|
3CFJ
 
 | |
3CFE
 
 | | Crystal structure of purple-fluorescent antibody EP2-25C10 | | Descriptor: | GLYCEROL, PURPLE-FLUORESCENT ANTIBODY EP2-25C10-IGG2B HEAVY CHAIN, PURPLE-FLUORESCENT ANTIBODY EP2-25C10-KAPPA LIGHT CHAIN, ... | | Authors: | Debler, E.W, Heine, A, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3CFK
 
 | | Crystal structure of catalytic elimination antibody 34E4, triclinic crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CATALYTIC ANTIBODY FAB 34E4 HEAVY CHAIN,Uncharacterized protein, ... | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-04 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational isomerism can limit antibody catalysis.
J.Biol.Chem., 283, 2008
|
|
3CFD
 
 | | Purple-fluorescent antibody EP2-25C10 in complex with its stilbene hapten | | Descriptor: | 4-(4-STYRYL-PHENYLCARBAMOYL)-BUTYRIC ACID, GLYCEROL, PURPLE-FLUORESCENT ANTIBODY EP2-25C10-IGG2B HEAVY CHAIN, ... | | Authors: | Debler, E.W, Heine, A, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3CFC
 
 | | High-resolution structure of blue fluorescent antibody EP2-19G2 | | Descriptor: | BLUE FLUORESCENT ANTIBODY EP2-19G2-IGG2B HEAVY CHAIN, BLUE FLUORESCENT ANTIBODY EP2-19G2-KAPPA LIGHT CHAIN, GLYCEROL | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3CFB
 
 | |
3F3P
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21212 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F3F
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3FO2
 
 | |
3F3G
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P212121 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F7F
 
 | | Structure of Nup120 | | Descriptor: | MERCURY (II) ION, Nucleoporin NUP120 | | Authors: | Seo, H.S, Ma, Y, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-11-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FO1
 
 | |
3FMP
 
 | | Crystal structure of the nucleoporin Nup214 in complex with the DEAD-box helicase Ddx19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | Napetschnig, J, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
