2QM7
 
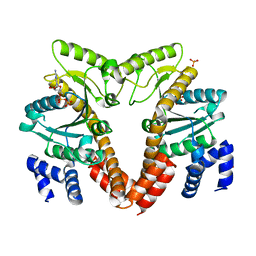 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP | | Descriptor: | GTPase/ATPase, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
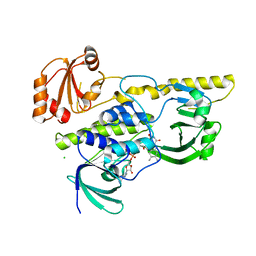
2R0C
 
 | |
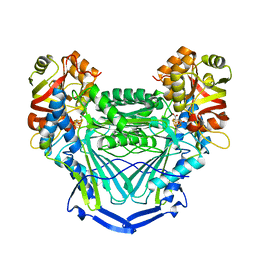
6MSN
 
 | |
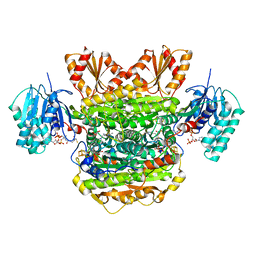
6N2O
 
 | |
6ND3
 
 | |
6NX0
 
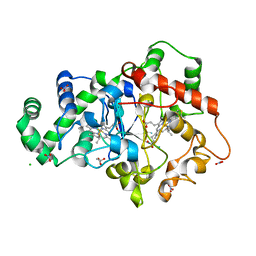 | |
6NHL
 
 | | Crystal structure of QueE from Escherichia coli | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Grell, T.A.J, Bell, B.N, Nguyen, C, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity.
Protein Sci., 28, 2019
|
|
6OND
 
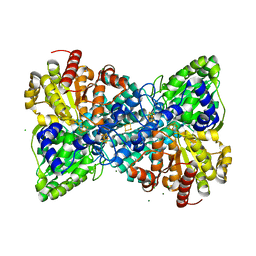 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase produced without CooC, reduced | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE (III) ION, ... | | Authors: | Wittenborn, E.C, Cohen, S.E, Drennan, C.L. | | Deposit date: | 2019-04-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6ONC
 
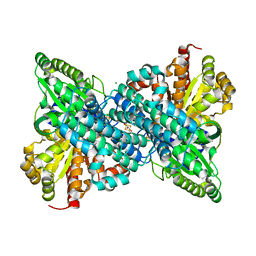 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase produced without CooC, as-isolated | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE (III) ION, ... | | Authors: | Wittenborn, E.C, Cohen, S.E, Drennan, C.L. | | Deposit date: | 2019-04-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6ONS
 
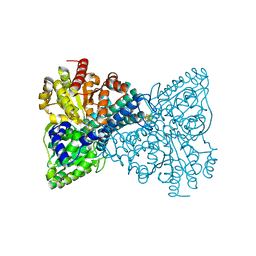 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase with the D-cluster ligating cysteines mutated to alanines, coexpressed with CooC, as-isolated | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Cohen, S.E, Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6OWR
 
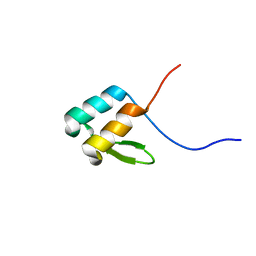 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
4K39
 
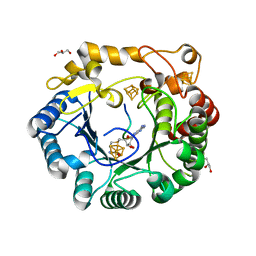 | | Native anSMEcpe with bound AdoMet and Cp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, Cp18Cys peptide, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M7S
 
 | |
4NJI
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Mg2+ | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJH
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxy-5,6,7,8-tetrahydropterin | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJJ
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Manganese(II) | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJG
 
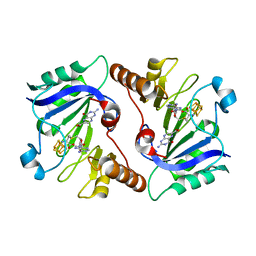 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxypterin | | Descriptor: | 6-CARBOXYPTERIN, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJK
 
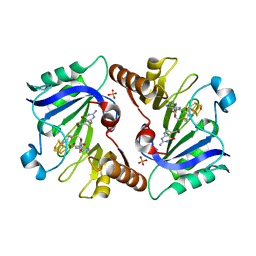 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4PKF
 
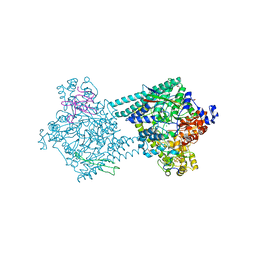 | | Benzylsuccinate synthase alpha-beta-gamma complex | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PKC
 
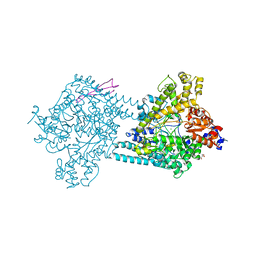 | | Benzylsuccinate alpha-gamma complex | | Descriptor: | CHLORIDE ION, GLYCEROL, TutD, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5T6O
 
 | |
5TK7
 
 | |
5T81
 
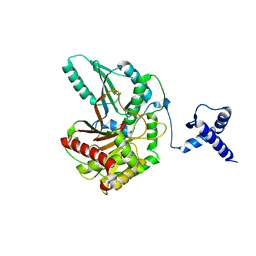 | | Rhombohedral crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB, GLYCEROL | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T8Y
 
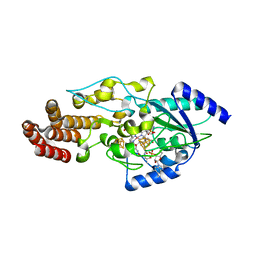 | | Structure of epoxyqueuosine reductase from Bacillus subtilis with the Asp134 catalytic loop swung out of the active site. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Maiocco, S.J, Elliott, S.J, Bandarian, V, Drennan, C.L. | | Deposit date: | 2016-09-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5TKA
 
 | |
