5DZT
 
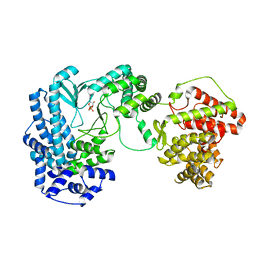 | |
4ZOQ
 
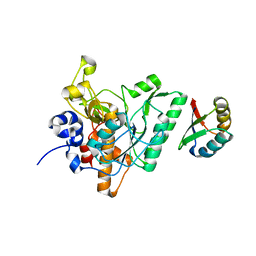 | | Crystal Structure of a Lanthipeptide Protease | | Descriptor: | Intracellular serine protease | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2015-05-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage.
Chem Sci, 6, 2015
|
|
6D6D
 
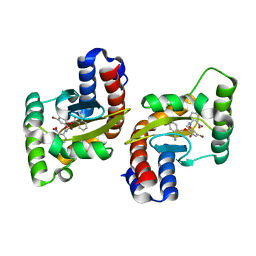 | |
6D6O
 
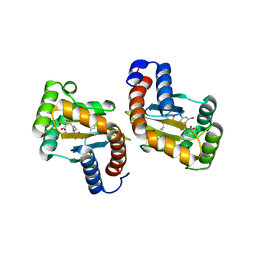 | |
6D6B
 
 | |
6D6C
 
 | |
6D6P
 
 | |
6D6L
 
 | |
6D6N
 
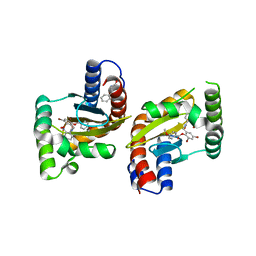 | | The structure of ligand binding domain of LasR in complex with TP-1 homolog, compound 16 | | Descriptor: | 2,4-dibromo-6-{[(2-nitrobenzene-1-carbonyl)amino]methyl}phenyl 4-methoxybenzoate, PHENYLALANINE, Transcriptional activator protein LasR | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop "Out" Conformation for LasR.
Cell Chem Biol, 25, 2018
|
|
6D6A
 
 | |
6D6M
 
 | |
6WNS
 
 | |
6WN0
 
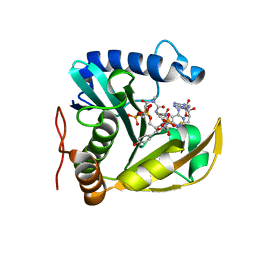 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
6OM4
 
 | |
6PEU
 
 | |
6PE3
 
 | |
5W8C
 
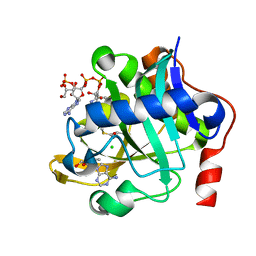 | | The structure of a COA-dependent acyl-homoserine lactone synthase, BjaI, with MTA and isovaleryl-CoA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Autoinducer synthase, CHLORIDE ION, ... | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2017-06-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the substrate specificity of quorum signal synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W8D
 
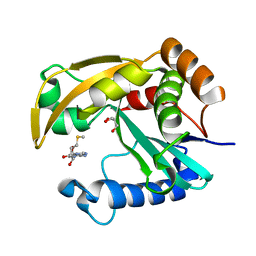 | |
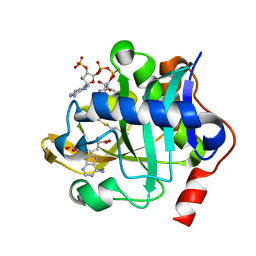
5W8A
 
 | |
5W8G
 
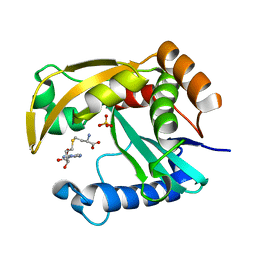 | |
5W8E
 
 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, BjaI, with the adduct of SAH and IV-CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-[(3-methylbutanoyl)amino]butanoic acid, ADENINE, Autoinducer synthase, ... | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2017-06-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the substrate specificity of quorum signal synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6XP8
 
 | |
5TV8
 
 | | A. aeolicus BioW with AMP-CPP and pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TVA
 
 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TV6
 
 | | A. aeolicus BioW with pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, PIMELIC ACID | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
