8BAW
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - 5-LICAM siderophore analogue complex. | | Descriptor: | FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Booth, R, Dodson, E.J, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BJ9
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - 5LICAM complex. | | Descriptor: | ABC transporter, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), ... | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BNW
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - apo form | | Descriptor: | ABC transporter, NICKEL (II) ION, SULFATE ION | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BF6
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - azotochelin complex | | Descriptor: | ABC transporter, Azotochelin, FE (III) ION, ... | | Authors: | Wilson, K.S, Duhme-Klair, A.-K, Blagova, E.V, Miller, A, Booth, R, Dodson, E.J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1RGP
 
 | | GTPASE-ACTIVATION DOMAIN FROM RHOGAP | | Descriptor: | RHOGAP | | Authors: | Barrett, T, Xiao, B, Dodson, E.J, Dodson, G, Ludbrook, S.B, Nurmahomed, K, Gamblin, S.J, Musacchio, A, Smerdon, S.J, Eccleston, J.F. | | Deposit date: | 1996-12-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the GTPase-activating domain from p50rhoGAP.
Nature, 385, 1997
|
|
1SAR
 
 | |
1CID
 
 | |
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
2BO4
 
 | | Dissection of mannosylglycerate synthase: an archetypal mannosyltransferase | | Descriptor: | CITRATE ANION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BO8
 
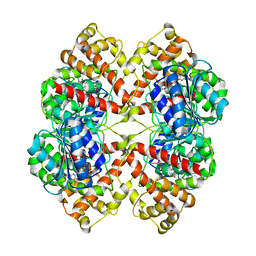 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
2BO6
 
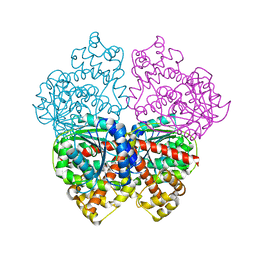 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, MANGANESE (II) ION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BO7
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
2BSX
 
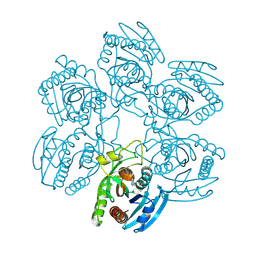 | | Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Schnick, C, Brzozowski, A.M, Dodson, E.J, Murshudov, G.N, Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Plasmodium Falciparum Purine Nucleoside Phosphorylase Complexed with Sulfate and its Natural Substrate Inosine
Acta Crystallogr.,Sect.D, 61, 2005
|
|
182D
 
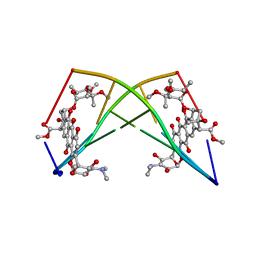 | | DNA-NOGALAMYCIN INTERACTIONS: THE CRYSTAL STRUCTURE OF D(TGATCA) COMPLEXED WITH NOGALAMYCIN | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Smith, C.K, Davies, G.J, Dodson, E.J, Moore, M.H. | | Deposit date: | 1994-07-28 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin.
Biochemistry, 34, 1995
|
|
3MIW
 
 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3QQ6
 
 | | The N-terminal DNA binding domain of SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator sinR | | Authors: | Colledge, V, Fogg, M.J, Levdikov, V.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2011-02-15 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Organisation of SinR, the Master Regulator of Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 411, 2011
|
|
3D55
 
 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
3CTO
 
 | | Crystal Structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
1I7O
 
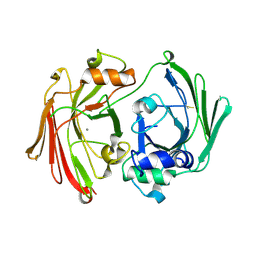 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-03-28 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of HpcE, a multi-functional enzyme fold
To be Published
|
|
1EB6
 
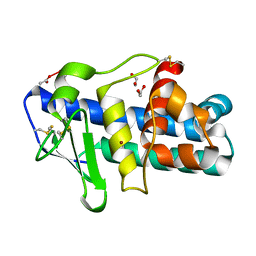 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FSE
 
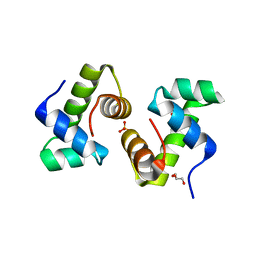 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
1LDN
 
 | | STRUCTURE OF A TERNARY COMPLEX OF AN ALLOSTERIC LACTATE DEHYDROGENASE FROM BACILLUS STEAROTHERMOPHILUS AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wigley, D.B, Gamblin, S.J, Turkenburg, J.P, Dodson, E.J, Piontek, K, Muirhead, H, Holbrook, J.J. | | Deposit date: | 1991-11-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution.
J.Mol.Biol., 223, 1992
|
|
1GTT
 
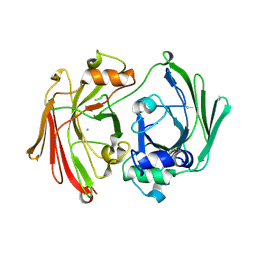 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2002-01-18 | | Release date: | 2002-03-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Biochemistry, 41, 2002
|
|
1OGK
 
 | | The crystal structure of Trypanosoma cruzi dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE, DEOXYURIDINE-5'-DIPHOSPHATE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1OGL
 
 | | The crystal structure of native Trypanosoma cruzi dUTPase | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
