1KR0
 
 | | Hevamine Mutant D125A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1KR1
 
 | | Hevamine Mutant D125A/E127A in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
2GDC
 
 | | Structure of Vinculin VD1 / IpaA560-633 complex | | Descriptor: | Invasin ipaA, Vinculin | | Authors: | Hamiaux, C, van Eerde, A, Parsot, C, Broos, J, Dijkstra, B.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri.
Embo Rep., 7, 2006
|
|
2GVY
 
 | |
2HTN
 
 | | E. coli bacterioferritin in its as-isolated form | | Descriptor: | Bacterioferritin, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | van Eerde, A, Wolterink-Van Loo, S, Van Der Oost, J, Dijkstra, B.W. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fortuitous structure determination of 'as-isolated' Escherichia coli bacterioferritin in a novel crystal form.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1CXH
 
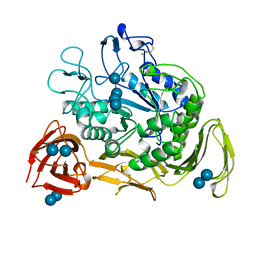 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.6 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH MALTOHEPTAOSE | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-31 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1CXE
 
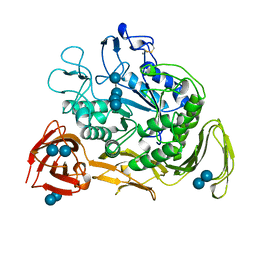 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.1 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1YOB
 
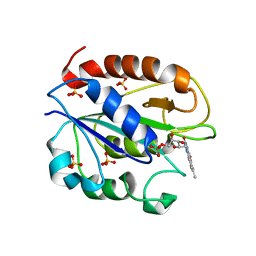 | | C69A Flavodoxin II from Azotobacter vinelandii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin 2, SULFATE ION | | Authors: | Alagaratnam, S, van Pouderoyen, G, Pijning, T, Dijkstra, B.W, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2005-01-27 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: structural determinants of redox potential
Protein Sci., 14, 2005
|
|
1CIU
 
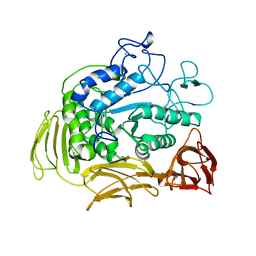 | |
1D3C
 
 | | MICHAELIS COMPLEX OF BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE WITH GAMMA-CYCLODEXTRIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, van der Veen, B.A, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 1999-09-29 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a gamma-cyclodextrin-CGTase complex at 1.8-A resolution.
J.Biol.Chem., 274, 1999
|
|
1CXI
 
 | |
1CXF
 
 | | COMPLEX OF A (D229N/E257Q) DOUBLE MUTANT CGTASE FROM BACILLUS CIRCULANS STRAIN 251 WITH MALTOTETRAOSE AT 120 K AND PH 9.1 OBTAINED AFTER SOAKING THE CRYSTAL WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1ZMT
 
 | | Structure of haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (R)-para-nitro styrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (R)-PARA-NITROSTYRENE OXIDE, Haloalcohol dehalogenase HheC | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Villa, A, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | Descriptor: | halohydrin dehalogenase | | Authors: | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
1ZO8
 
 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
2AE0
 
 | | Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Membrane-bound lytic murein transglycosylase A | | Authors: | Van Straaten, K.E, Dijkstra, B.W, Vollmer, W, Thunnissen, A.M.W.H. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of MltA from Escherichia coli Reveals a Unique Lytic Transglycosylase Fold
J.Mol.Biol., 352, 2005
|
|
2B4K
 
 | | Acetobacter turbidans alpha-amino acid ester hydrolase complexed with phenylglycine | | Descriptor: | Alpha-amino acid ester hydrolase, D-PHENYLGLYCINE, GLYCEROL | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Williams, C, Wybenga, G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
1QD5
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
1QD6
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CALCIUM ION, OUTER MEMBRANE PHOSPHOLIPASE (OMPLA), ... | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
1SKZ
 
 | | PROTEASE INHIBITOR | | Descriptor: | ANTISTASIN, CHLORIDE ION | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa.
EMBO J., 16, 1997
|
|
1UKR
 
 | | STRUCTURE OF ENDO-1,4-BETA-XYLANASE C | | Descriptor: | ENDO-1,4-B-XYLANASE I | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1996-08-23 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
J.Mol.Biol., 263, 1996
|
|
1EX9
 
 | | CRYSTAL STRUCTURE OF THE PSEUDOMONAS AERUGINOSA LIPASE COMPLEXED WITH RC-(RP,SP)-1,2-DIOCTYLCARBAMOYL-GLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, LACTONIZING LIPASE, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER | | Authors: | Nardini, M, Lang, D.A, Liebeton, K, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2000-05-02 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases.
J.Biol.Chem., 275, 2000
|
|
1FP9
 
 | |
1FP8
 
 | | STRUCTURE OF THE AMYLOMALTASE FROM THERMUS THERMOPHILUS HB8 IN SPACE GROUP P21212 | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CHLORIDE ION, MERCURY (II) ION | | Authors: | Uitdehaag, J.C.M, Euverink, G.J, van der Veen, B.A, van der Maarel, M, Dijkstra, B.W. | | Deposit date: | 2000-08-31 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the amylomaltase from Thermus thermophilus HB8
To be Published
|
|
1FXH
 
 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
