1C9H
 
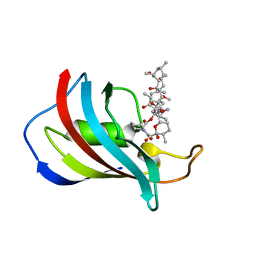 | | CRYSTAL STRUCTURE OF FKBP12.6 IN COMPLEX WITH RAPAMYCIN | | Descriptor: | FKBP12.6, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Deivanayagam, C.C.S, Carson, M, Thotakura, A, Narayana, S.V.L, Chodavarapu, C.S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FKBP12.6 in complex with rapamycin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1D2P
 
 | |
1D2O
 
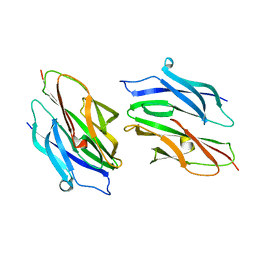 | |
1N67
 
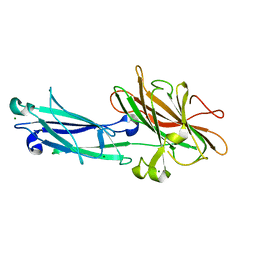 | | Clumping Factor A from Staphylococcus aureus | | Descriptor: | Clumping Factor, MAGNESIUM ION | | Authors: | Deivanayagam, C.C.S, Wann, E.R, Chen, W, Carson, M, Rajashankar, K.R, Hook, M, Narayana, S.V.L. | | Deposit date: | 2002-11-08 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel variant of the immunoglobulin fold in surface adhesins of
Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM,
clumping factor A
Embo J., 21, 2002
|
|
1QC5
 
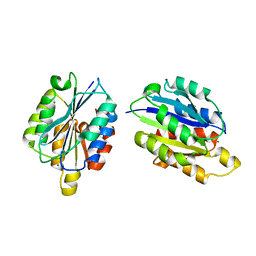 | | I Domain from Integrin Alpha1-Beta1 | | Descriptor: | MAGNESIUM ION, PROTEIN (ALPHA1 BETA1 INTEGRIN) | | Authors: | Deivanayagam, C.C, Narayana, S.V. | | Deposit date: | 1999-05-17 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM.
J.Biol.Chem., 274, 1999
|
|
3AU0
 
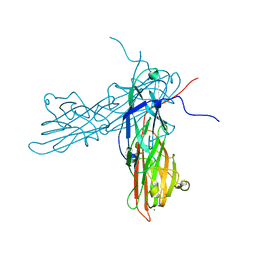 | | Structural and biochemical characterization of ClfB:ligand interactions | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Ganesh, V.K, Barbu, E.M, Deivanayagam, C.C.S, Le, B, Anderson, A.S, Matsuka, Y, Lin, S.L, Foster, T.F, Narayana, S.V.L, Hook, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biochemical characterization of ClfB:ligand interactions
To be published
|
|
1NX0
 
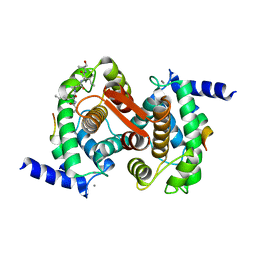 | | Structure of Calpain Domain 6 in Complex with Calpastatin DIC | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX1
 
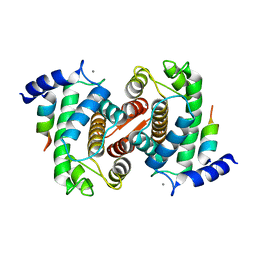 | | Calpain Domain VI Complexed with Calpastatin Inhibitory Domain C (DIC) | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX3
 
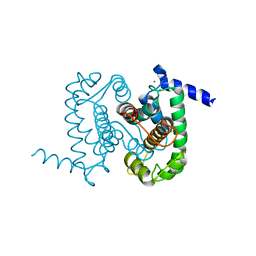 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX2
 
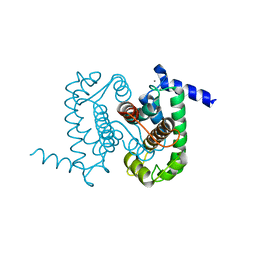 | | Calpain Domain VI | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
3IOX
 
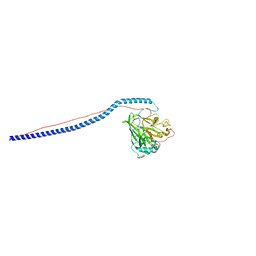 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFITE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IPK
 
 | | Crystal Structure of A3VP1 of AgI/II of Streptococcus mutans | | Descriptor: | AgI/II, CALCIUM ION, SULFATE ION, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Patel, M, Robinette, R, Crowley, P, Michalek, S.M, Brady, L.J, Deivanayagam, C.C. | | Deposit date: | 2009-08-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DF0
 
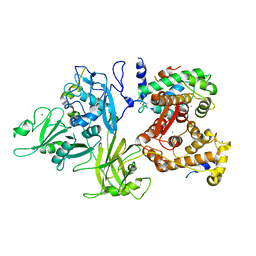 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
6MEN
 
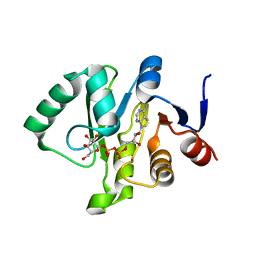 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEB
 
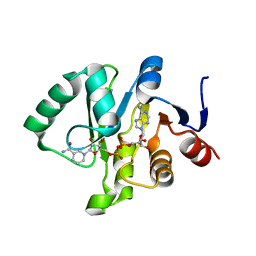 | | Crystal structure of Tylonycteris bat coronavirus HKU4 macrodomain in complex with nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEA
 
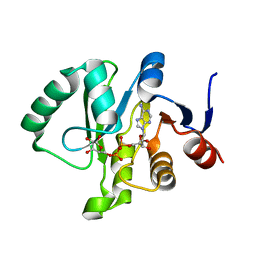 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
3ASW
 
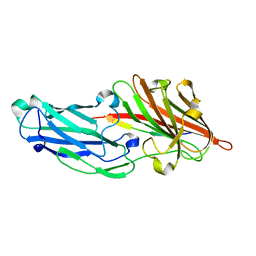 | |
3AT0
 
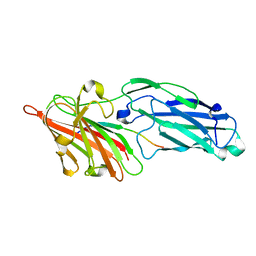 | |
