8B5L
 
 | | Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
8B6C
 
 | | Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147 | | Descriptor: | 28S rRNA, 4-[[9-(cyclohexylmethyl)-3-methylidene-5-(4-methylphenyl)sulfonyl-1,5,9-triazacyclododec-1-yl]sulfonyl]-~{N},~{N}-dimethyl-aniline, 5.8S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
6YMV
 
 | |
6YMW
 
 | |
8Q63
 
 | | Cryo-EM structure of IC8', a second state of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8AP1
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with two GTP molecules poised for de novo initiation (IC2) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATV
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C5U
 
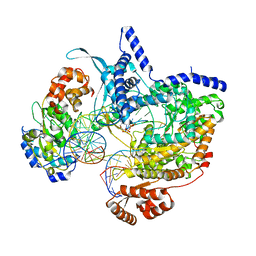 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C5S
 
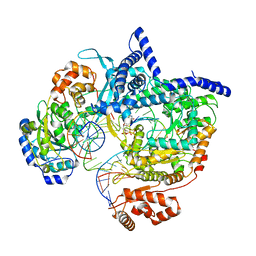 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
7Z2H
 
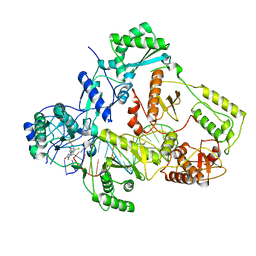 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z24
 
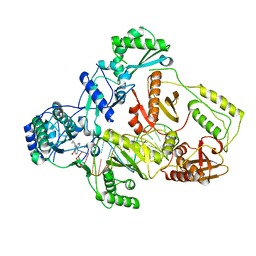 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-mer), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2G
 
 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2D
 
 | |
7Z29
 
 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2E
 
 | |
