3ONE
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3PJB
 
 | |
3PIB
 
 | |
3PJ7
 
 | |
3PJ5
 
 | |
1XYZ
 
 | |
1SVN
 
 | | SAVINASE | | Descriptor: | CALCIUM ION, SAVINASE (TM) | | Authors: | Betzel, C, Klupsch, S, Papendorf, G, Hastrup, S, Branner, S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the alkaline proteinase Savinase from Bacillus lentus at 1.4 A resolution.
J.Mol.Biol., 223, 1992
|
|
5TOV
 
 | |
5TOW
 
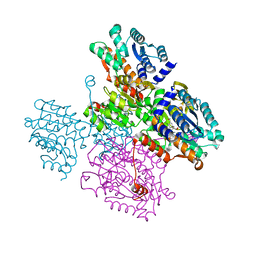 | | Crystal structure of the inactive form of S-adenosyl-L-homocysteine hydrolase from Thermotoga maritima in ternary complex with NADH and Adenosine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE, ... | | Authors: | Czyrko, J, Brzezinski, K. | | Deposit date: | 2016-10-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S-adenosyl-L-homocysteine hydrolase from a hyperthermophile (Thermotoga maritima) is expressed in Escherichia coli in inactive form - Biochemical and structural studies.
Int. J. Biol. Macromol., 104, 2017
|
|
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1C54
 
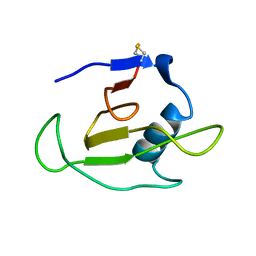 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-14 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
5T3F
 
 | |
5T3J
 
 | |
5T3G
 
 | | thaumatin soaked with selenourea for 10 min | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1, selenourea | | Authors: | Luo, Z, Dauter, Z. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selenourea: a convenient phasing vehicle for macromolecular X-ray crystal structures.
Sci Rep, 6, 2016
|
|
5T3I
 
 | |
5T3H
 
 | | bovine trypsin soaked with selenourea for 5 min | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Luo, Z, Dauter, Z. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selenourea: a convenient phasing vehicle for macromolecular X-ray crystal structures.
Sci Rep, 6, 2016
|
|
5T3L
 
 | | B-DNA (CGCGAATTCGCG)2 soaked with selenourea for 1 min | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), selenourea | | Authors: | Luo, Z, Dauter, Z. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Selenourea: a convenient phasing vehicle for macromolecular X-ray crystal structures.
Sci Rep, 6, 2016
|
|
6M9Z
 
 | |
6M9Y
 
 | |
6MAS
 
 | | X-ray Structure of Branchiostoma floridae fluorescent protein lanFP10G | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V, Pletnev, S. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Factors Enabling Successful GFP-Like Proteins with Alanine as the Third Chromophore-Forming Residue.
J. Mol. Biol., 431, 2019
|
|
