4J3B
 
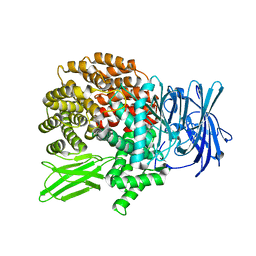 | | A naturally variable residue in the S1 subsite of M1-family aminopeptidases modulates catalytic properties and promotes functional specialization | | Descriptor: | ARGININE, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Dalal, S, Ragheb, D.R.T, Schubot, F.D, Klemba, M. | | Deposit date: | 2013-02-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A naturally variable residue in the s1 subsite of m1 family aminopeptidases modulates catalytic properties and promotes functional specialization.
J.Biol.Chem., 288, 2013
|
|
6QPP
 
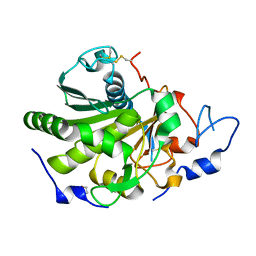 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
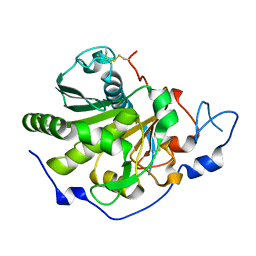 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
3V7J
 
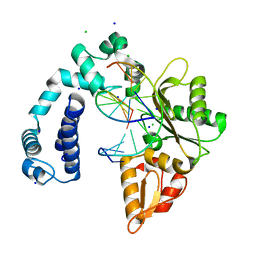 | | Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*AP*AP*CP*TP*CP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3UXO
 
 | |
3Q43
 
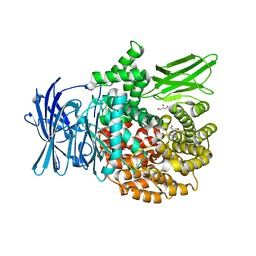 | |
3Q44
 
 | |
3T8V
 
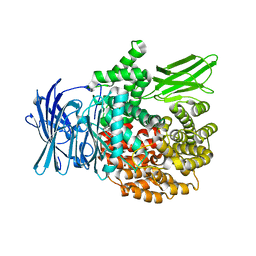 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | M1 family aminopeptidase, MAGNESIUM ION, N-[(2-{2-[(N-{(2S,3R)-3-amino-4-[4-(benzyloxy)phenyl]-2-hydroxybutanoyl}-L-alanyl)amino]ethoxy}ethoxy)acetyl]-4-benzoyl-L-phenylalanyl-N~6~-hex-5-ynoyllysinamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T8W
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-((2R,3S,6S,18S,21S)-2-amino-18-(4-benzoylbenzyl)-21-carbamoyl-3-hydroxy-6-(naphthalen-2-ylmethyl)-4,7,16,19-tetraoxo-1-phenyl-11,14-dioxa-5,8,17,20-tetraazapentacosan-25-yl)hex-5-ynamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3V7K
 
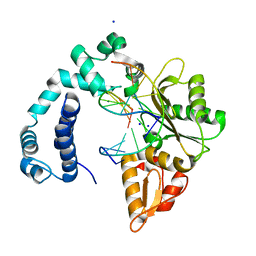 | |
3V7L
 
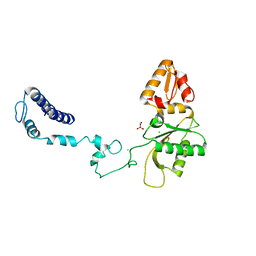 | | Apo Structure of Rat DNA polymerase beta K72E variant | | Descriptor: | CHLORIDE ION, DNA polymerase beta, SODIUM ION, ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3UXN
 
 | |
3UXP
 
 | | Co-crystal Structure of Rat DNA polymerase beta Mutator I260Q: Enzyme-DNA-ddTTP | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA 5'-D(P*AP*CP*TP*CP*AP*CP*AP*TP*A)-3', DNA 5'-D(P*AP*TP*GP*TP*GP*AP*G)-3', ... | | Authors: | Gridley, C.L, Jaeger, J. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural Changes in the Hydrophobic Hinge Region Adversely Affect the Activity and Fidelity of the I260Q Mutator DNA Polymerase beta.
Biochemistry, 52, 2013
|
|
