6BBR
 
 | |
6BC2
 
 | |
6BC7
 
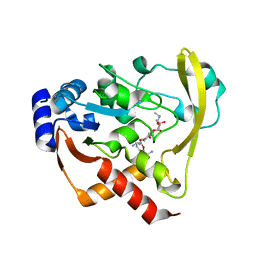 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, ACETATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC6
 
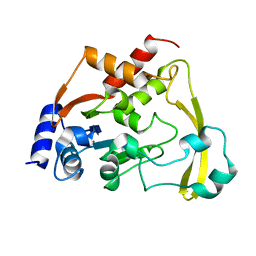 | | Cryo X-ray structure of apo AAC-VIa | | Descriptor: | AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC3
 
 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, COENZYME A | | Authors: | Cuneo, M.J, Kumar, P, Serpersu, E.H. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC5
 
 | |
2GX6
 
 | |
6NMK
 
 | |
6NMM
 
 | |
6NTI
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NLT
 
 | | Apo structure of the T130K mutant of ANT-4 | | Descriptor: | CALCIUM ION, Kanamycin nucleotidyltransferase | | Authors: | Selvaraj, B, Cuneo, M.J. | | Deposit date: | 2019-01-09 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catch and release: A novel variation of the archetypal nucleotidyl transfer reaction
To be published
|
|
6NML
 
 | |
6NTJ
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NMN
 
 | |
5HM4
 
 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
5H8Z
 
 | |
4JSO
 
 | |
4JSD
 
 | |
4KSE
 
 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
3MQ1
 
 | | Crystal Structure of Dust Mite Allergen Der p 5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
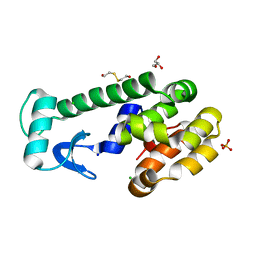 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
6MEZ
 
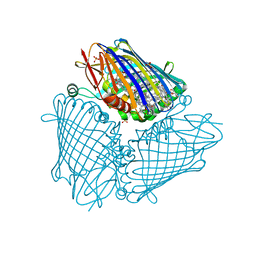 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | Authors: | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
4PFU
 
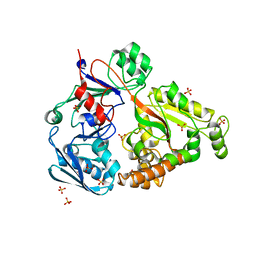 | | Crystal structure of mannobiose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.05 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
4P1H
 
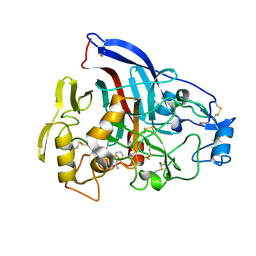 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, Exoglucanase 1, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form
to be published
|
|
