3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3EY8
 
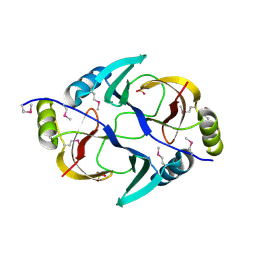 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
3L9A
 
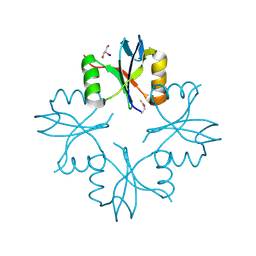 | | Structure of the C-terminal domain from a Streptococcus mutans hypothetical | | Descriptor: | GLYCEROL, SODIUM ION, uncharacterized protein | | Authors: | Singer, A.U, Cuff, M.E, Xu, X, Cui, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-04 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the C-terminal domain from a Streptococcus mutans hypothetical
To be Published
|
|
3LED
 
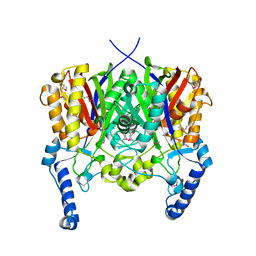 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009 | | Descriptor: | 3-oxoacyl-acyl carrier protein synthase III, FORMIC ACID | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009
To be Published
|
|
3F4A
 
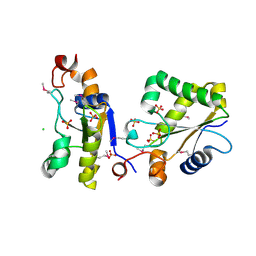 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3F5R
 
 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
3LEQ
 
 | | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A | | Descriptor: | uncharacterized protein cvnB5 | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A
To be Published
|
|
3LL4
 
 | | Structure of the H13A mutant of Ykr043C in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Stogios, P.J, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3EY7
 
 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, CALCIUM ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1
To be Published
|
|
3FHM
 
 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
3FBU
 
 | | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, COENZYME A | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis
To be Published
|
|
3LG2
 
 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | Descriptor: | PHOSPHATE ION, Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3LVY
 
 | | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Carboxymuconolactone decarboxylase family, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans
To be Published
|
|
3LZK
 
 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3MQZ
 
 | | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA. | | Descriptor: | CHLORIDE ION, GLYCEROL, uncharacterized Conserved Protein DUF1054 | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA.
To be Published
|
|
3M6J
 
 | | Crystal structure of unknown function protein from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-03-31 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of unknown function protein from Leptospirillum rubarum
To be Published
|
|
3MAJ
 
 | | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris CGA009 | | Descriptor: | DNA processing chain A, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris
To be Published
|
|
3G1J
 
 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4. | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
To be published
|
|
3G64
 
 | | Crystal structure of putative enoyl-CoA hydratase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative enoyl-CoA hydratase, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Putative Enoyl-CoA Hydratase from Streptomyces coelicolor A3(2)
To be Published
|
|
3M92
 
 | | The structure of yciN, an unchracterized protein from Shigella flexneri. | | Descriptor: | CHLORIDE ION, Protein yciN, SODIUM ION | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-19 | | Release date: | 2010-05-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of yciN, an unchracterized protein from Shigella flexneri.
TO BE PUBLISHED
|
|
3MFN
 
 | | Dfer_2879 protein of unknown function from Dyadobacter fermentans | | Descriptor: | ACETATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Chin, S, Eisen, J, Wu, D, Kerfeld, C, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | X-ray crystal structure of Dfer_2879 protein of unknown function from Dyadobacter fermentans.
To be Published
|
|
3GEM
 
 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
3GK6
 
 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
3MTJ
 
 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
