4E27
 
 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences Solved by Iodide SAD Phasing | | Descriptor: | Capsid Protein, IODIDE ION, SODIUM ION | | Authors: | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences Solved by Iodide SAD Phasing
To be Published
|
|
3VFI
 
 | | Crystal Structure of a Metagenomic Thioredoxin | | Descriptor: | thioredoxin | | Authors: | Craig, T.K, Gardberg, A, Lorimer, D.D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Metagenomic Thioredoxin
To be Published
|
|
4G7F
 
 | |
4DMI
 
 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences (CASP) | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, SODIUM ION | | Authors: | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-02-07 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences
To be Published
|
|
3N5A
 
 | | Synaptotagmin-7, C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Craig, T.K. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7.
Plos One, 5, 2010
|
|
4F2G
 
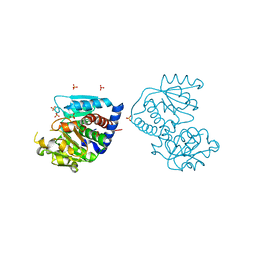 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | Authors: | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4H4G
 
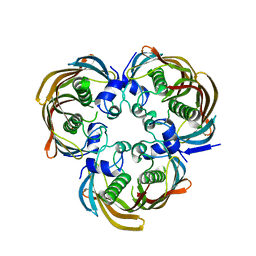 | | Crystal Structure of (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase from Burkholderia thailandensis E264 | | Descriptor: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | Authors: | Craig, T.K, Edwards, T.E, Staker, B, Stewart, L, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4CSO
 
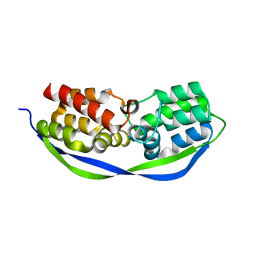 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
2M7T
 
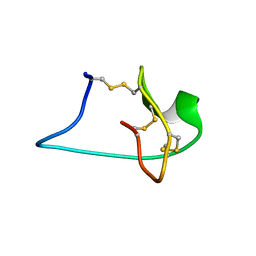 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
4HJH
 
 | |
4EIT
 
 | |
4EFZ
 
 | |
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUL
 
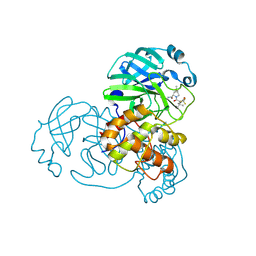 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
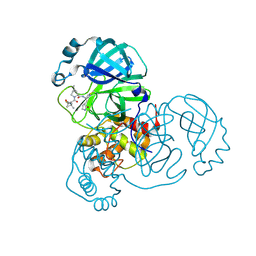 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
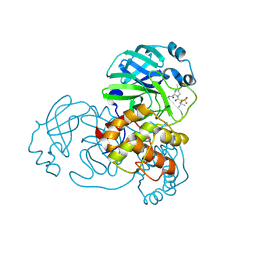 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
6ANJ
 
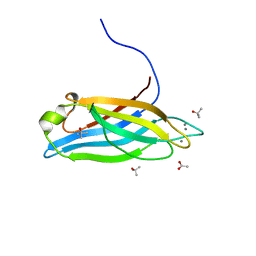 | | Synaptotagmin-7, C2A domain | | Descriptor: | ACETATE ION, CALCIUM ION, Synaptotagmin-7, ... | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ANK
 
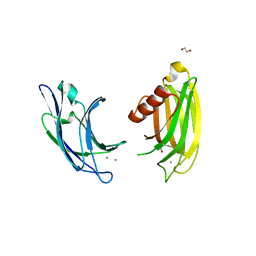 | | Synaptotagmin-7, C2A- and C2B-domains | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7OFV
 
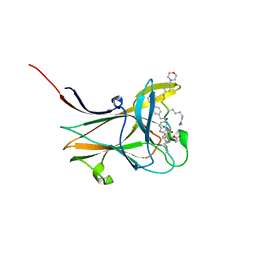 | | NMR-guided design of potent and selective EphA4 agonistic ligands | | Descriptor: | ACETATE ION, EphA4 agonist ligand, Ephrin type-A receptor 4 | | Authors: | Ganichkin, O.M, Craig, T.K, Baggio, C, Pellecchia, M. | | Deposit date: | 2021-05-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | NMR-Guided Design of Potent and Selective EphA4 Agonistic Ligands.
J.Med.Chem., 64, 2021
|
|
4EFI
 
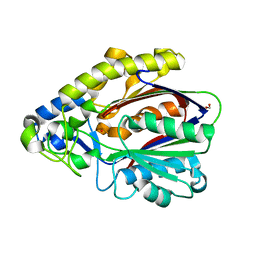 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | Descriptor: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
