7ZRM
 
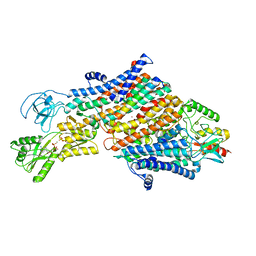 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
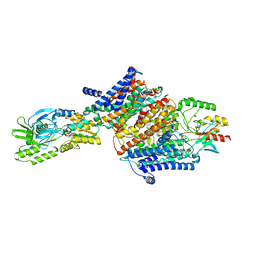 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
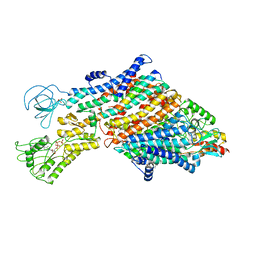 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
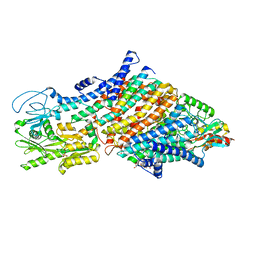 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRD
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
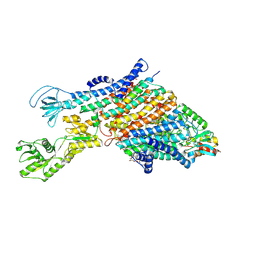 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7NNP
 
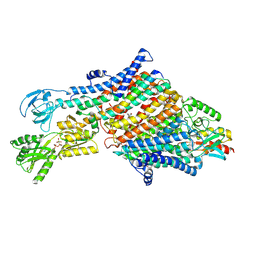 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNL
 
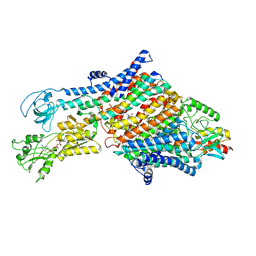 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
8B70
 
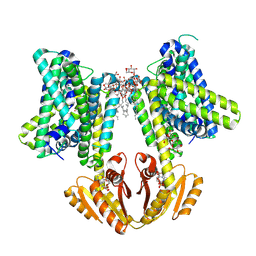 | | KimA from B. subtilis with nucleotide second-messenger c-di-AMP bound | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
8B71
 
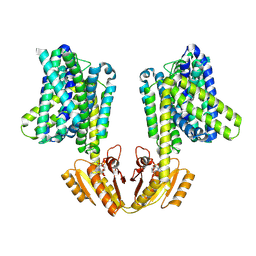 | | Upright KimA dimer with bound c-di-AMP from B. subtilis | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Potassium transporter KimA | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
8CH5
 
 | |
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
7NY1
 
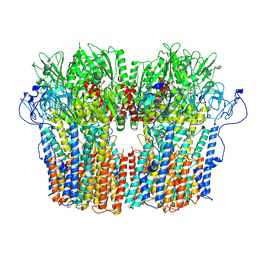 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NXF
 
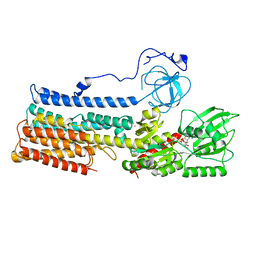 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
6M96
 
 | |
6S3K
 
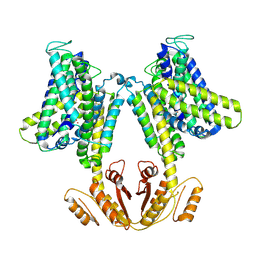 | | KimA from Bacillus subtilis in inward-facing, occluded state | | Descriptor: | APC family permease, POTASSIUM ION | | Authors: | Tascon, I, Sousa, J.S, Vonck, J, Haenelt, I. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of proton-coupled potassium transport in the KUP family.
Nat Commun, 11, 2020
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
8TLU
 
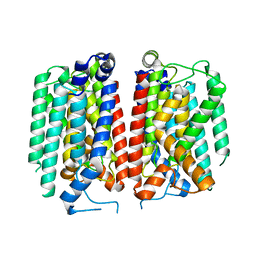 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
6QSK
 
 | |
6R7X
 
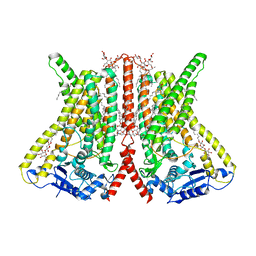 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
7OXR
 
 | |
