7VX1
 
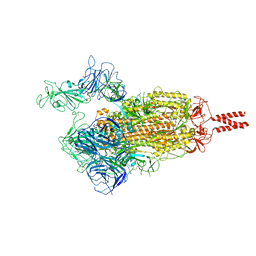 | | SARS-CoV-2 Beta variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXE
 
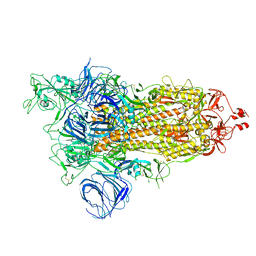 | | SARS-CoV-2 Kappa variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXF
 
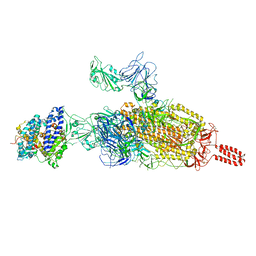 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2B state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXD
 
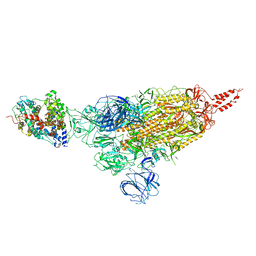 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C1 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX4
 
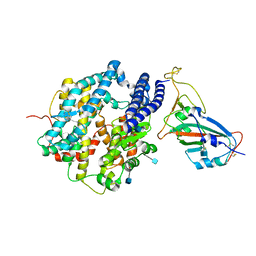 | | ACE2-RBD in SARS-CoV-2 Beta variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX5
 
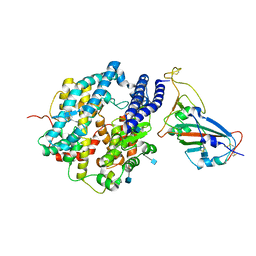 | | ACE2-RBD in SARS-CoV-2 Kappa variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXK
 
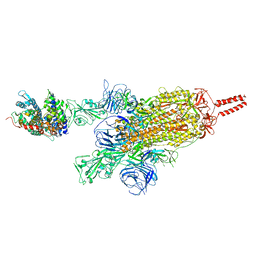 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2A state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXI
 
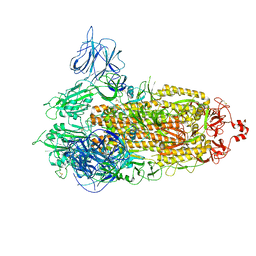 | | SARS-CoV-2 Kappa variant spike protein in transition state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXB
 
 | | SARS-CoV-2 Kappa variant spike protein in C2b state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXC
 
 | | SARS-CoV-2 Kappa variant spike protein in C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXM
 
 | |
5YHQ
 
 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
5WVI
 
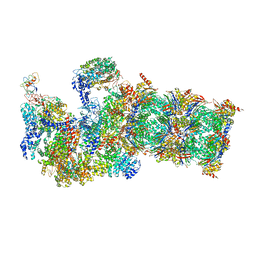 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5WVK
 
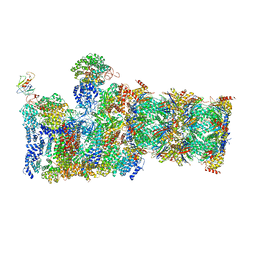 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
8X91
 
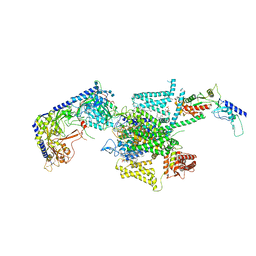 | | P/Q type calcium channel in complex with omega-conotoxin MVIIC | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 2024
|
|
8X93
 
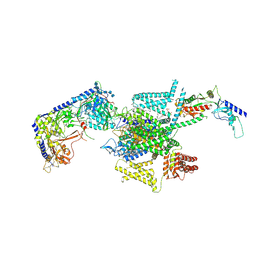 | | P/Q type calcium channel in complex with omega-Agatoxin IVA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 2024
|
|
8X90
 
 | | P/Q type calcium channel | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 2024
|
|
7V5M
 
 | | 20S+monoUb-CyclinB1-NT (S2) | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
7V5G
 
 | | 20S+monoUb-CyclinB1-NT (S1) | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
5TSL
 
 | |
5TSK
 
 | |
6KRE
 
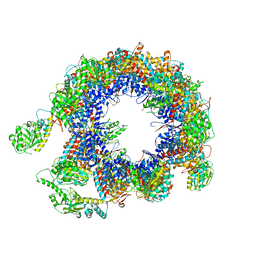 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
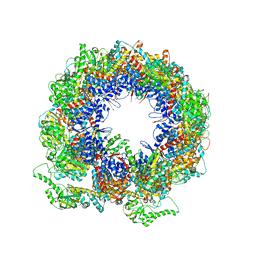 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
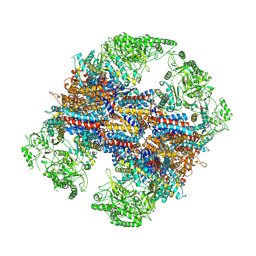 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
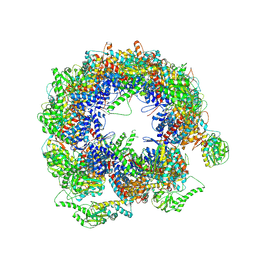 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
