2FK6
 
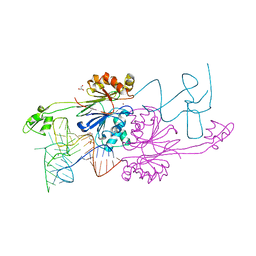 | | Crystal Structure of RNAse Z/tRNA(Thr) complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, RIBONUCLEASE Z, ... | | 著者 | Li de la Sierra-Gallay, I, Mathy, N, Pellegrini, O, Condon, C. | | 登録日 | 2006-01-04 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3T3N
 
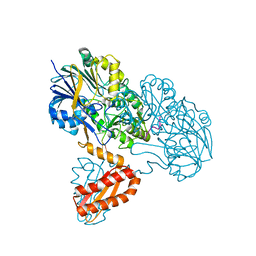 | | Molecular basis for the recognition and cleavage of RNA (UUCCGU) by the bifunctional 5'-3' exo/endoribonuclease RNase J | | 分子名称: | Metal dependent hydrolase, O2'methyl-RNA, ZINC ION | | 著者 | Dorleans, A, Li de la Sierra-Gallay, I, Piton, J, Zig, L, Gilet, L, Putzer, H, Condon, C. | | 登録日 | 2011-07-25 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Molecular Basis for the Recognition and Cleavage of RNA by the Bifunctional 5'-3' Exo/Endoribonuclease RNase J.
Structure, 19, 2011
|
|
3T3O
 
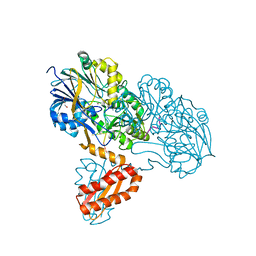 | | Molecular basis for the recognition and cleavage of RNA (CUGG) by the bifunctional 5'-3' exo/endoribonuclease RNase J | | 分子名称: | GLYCEROL, Metal dependent hydrolase, O2'methyl-RNA, ... | | 著者 | Dorleans, A, Li de la Sierra-Gallay, I, Piton, J, Zig, L, Gilet, L, Putzer, H, Condon, C. | | 登録日 | 2011-07-25 | | 公開日 | 2011-10-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular Basis for the Recognition and Cleavage of RNA by the Bifunctional 5'-3' Exo/Endoribonuclease RNase J.
Structure, 19, 2011
|
|
1Y44
 
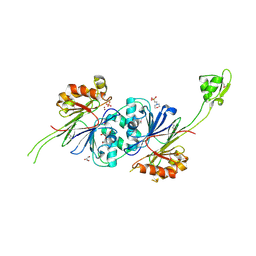 | | Crystal structure of RNase Z | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | 著者 | de la Sierra-Gallay, I.L, Pellegrini, O, Condon, C. | | 登録日 | 2004-11-30 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature, 433, 2005
|
|
6TPQ
 
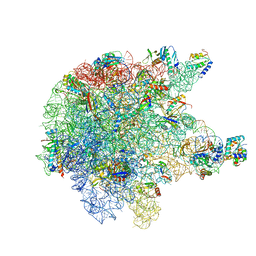 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TNN
 
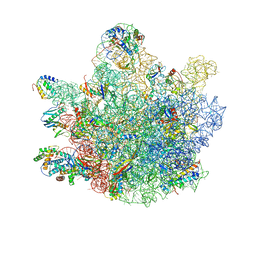 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-09 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
5MQ8
 
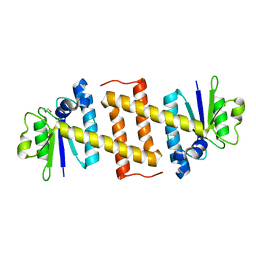 | | Crystal structure of Rae1 (YacP) from Bacillus subtilis | | 分子名称: | GLYCEROL, Uncharacterized protein YacP | | 著者 | Piton, J, Gilet, L, Pellegrini, O, Leroy, M, Figaro, S, Condon, C. | | 登録日 | 2016-12-20 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Rae1/YacP, a new endoribonuclease involved in ribosome-dependent mRNA decay in Bacillus subtilis.
EMBO J., 36, 2017
|
|
5MQ9
 
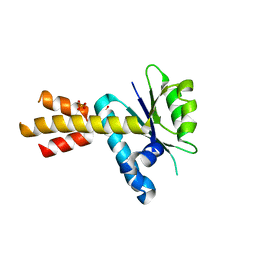 | | Crystal structure of Rae1 (YacP) from Bacillus subtilis (W164L mutant) | | 分子名称: | SULFATE ION, Uncharacterized protein YacP | | 著者 | Piton, J, Gilet, L, Pellegrini, O, Leroy, M, Figaro, S, Condon, C. | | 登録日 | 2016-12-20 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.174 Å) | | 主引用文献 | Rae1/YacP, a new endoribonuclease involved in ribosome-dependent mRNA decay in Bacillus subtilis.
EMBO J., 36, 2017
|
|
4GCW
 
 | | Crystal structure of RNase Z in complex with precursor tRNA(Thr) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ribonuclease Z, TRNA(THR), ... | | 著者 | Pellegrini, O, Li de la Sierra-Gallay, I, Piton, J, Gilet, L, Condon, C. | | 登録日 | 2012-07-31 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Activation of tRNA Maturation by Downstream Uracil Residues in B. subtilis
Structure, 20, 2012
|
|
4JZT
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZS
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6Z2B
 
 | |
6Y5U
 
 | |
6Y56
 
 | |
5DPX
 
 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | 分子名称: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | 著者 | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | 登録日 | 2015-09-14 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
6TGJ
 
 | |
6TG6
 
 | | Toprim domain of RNase M5 | | 分子名称: | Ribonuclease M5 | | 著者 | Oerum, S, Catala, M, Tisne, C. | | 登録日 | 2019-11-15 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | 分子名称: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | 著者 | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | 登録日 | 2017-01-11 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
