2QHK
 
 | |
2QM3
 
 | | Crystal structure of a predicted methyltransferase from Pyrococcus furiosus | | Descriptor: | ACETIC ACID, CALCIUM ION, Predicted methyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a predicted methyltransferase from Pyrococcus furiosus.
TO BE PUBLISHED
|
|
2QSW
 
 | | Crystal structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis | | Descriptor: | GLYCEROL, Methionine import ATP-binding protein metN 2, ZINC ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis.
To be Published
|
|
2QLC
 
 | |
2R4Q
 
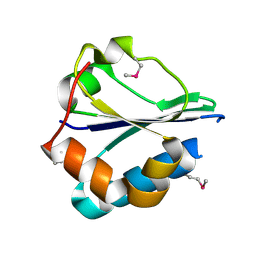 | | The structure of a domain of fruA from Bacillus subtilis | | Descriptor: | Phosphotransferase system (PTS) fructose-specific enzyme IIABC component | | Authors: | Cuff, M.E, Sather, A, Nocek, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a domain of fruA from Bacillus subtilis.
TO BE PUBLISHED
|
|
2R48
 
 | | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Phosphotransferase system (PTS) mannose-specific enzyme IIBCA component | | Authors: | Nocek, B, Cuff, M, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
2R78
 
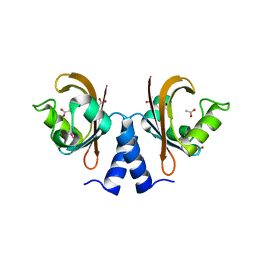 | |
2R5F
 
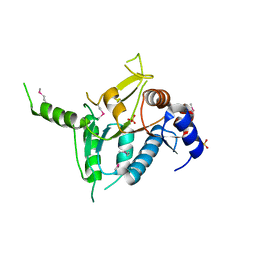 | | Putative sugar-binding domain of transcriptional regulator DeoR from Pseudomonas syringae pv. tomato | | Descriptor: | SULFATE ION, Transcriptional regulator, putative | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Putative sugar-binding domain of trancsriptional regulator DeoR from Pseudomonas syringae pv. tomato.
TO BE PUBLISHED
|
|
2RK5
 
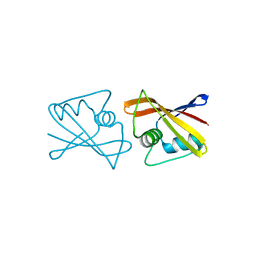 | |
4O2H
 
 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | Descriptor: | protein BCAM1869 | | Authors: | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-22 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
3OBF
 
 | | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302 | | Descriptor: | Putative transcriptional regulator, IclR family | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302
To be Published
|
|
3OJ0
 
 | | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain) | | Descriptor: | GLYCEROL, Glutamyl-tRNA reductase, SULFATE ION | | Authors: | Michalska, K, Marshall, N, Clancy, S, Puttagunta, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain)
To be Published
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3OVK
 
 | |
3OOS
 
 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
3PEB
 
 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
3PFM
 
 | | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf | | Descriptor: | GGDEF domain protein | | Authors: | Nocek, B, Stein, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf
TO BE PUBLISHED
|
|
3PU9
 
 | | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein serine/threonine phosphatase | | Authors: | Nocek, B, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745
TO BE PUBLISHED
|
|
3PN9
 
 | |
3QOO
 
 | | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans
TO BE PUBLISHED
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
3QZ6
 
 | |
3RNR
 
 | | Crystal Structure of Stage II Sporulation E Family Protein from Thermanaerovibrio acidaminovorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Stage II Sporulation E Family Protein from Thermanaerovibrio acidaminovorans
To be Published
|
|
3RCN
 
 | | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens | | Descriptor: | ACETIC ACID, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens
To be Published
|
|
