2XON
 
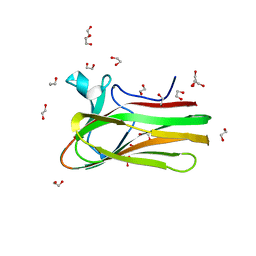 | | Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, ... | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XOM
 
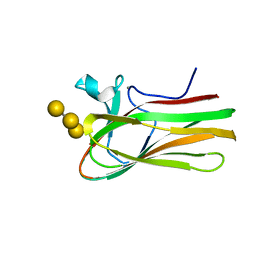 | | Atomic resolution structure of TmCBM61 in complex with beta-1,4- galactotriose | | Descriptor: | ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
6YT5
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly | | Descriptor: | Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YSZ
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly | | Descriptor: | Internal virion protein gp15 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3J4A
 
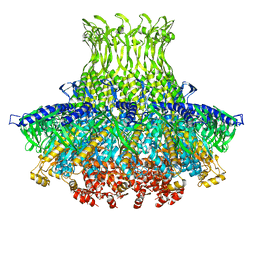 | | Structure of gp8 connector protein | | Descriptor: | Head-to-tail joining protein | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3J4B
 
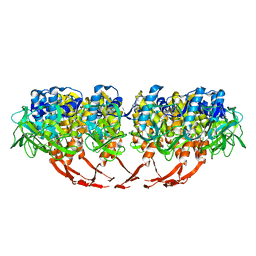 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
4BIJ
 
 | | Threading model of T7 large terminase | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
4BIL
 
 | | Threading model of the T7 large terminase within the gp8gp19 complex | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
4A41
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with galactose | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, SODIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A44
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylglucosaminidase, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4AAX
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-12-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A6O
 
 | | CpGH89CBM32-4, produced by Clostridium perfringens, in complex with glcNAc-alpha-1,4-galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A45
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A42
 
 | | CpGH89CBM32-6 produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A3Z
 
 | | CpGH89CBM32-4 (seleno-methionine labeled) produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
1GBR
 
 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
7Z38
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
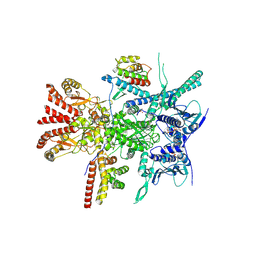 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
5A6J
 
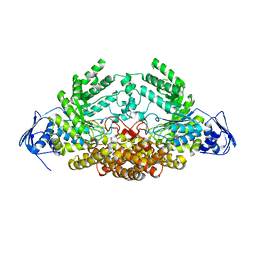 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A6K
 
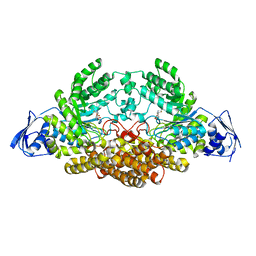 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-NGT | | Descriptor: | (2S,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, GH20C | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Second Beta-Hexosaminidase Encoded in the Streptococcus Pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
J.Biol.Chem., 290, 2015
|
|
5A6B
 
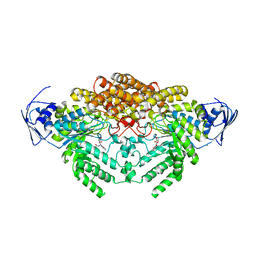 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with PUGNAc | | Descriptor: | MAGNESIUM ION, N-ACETYL-BETA-D-GLUCOSAMINIDASE, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5AC5
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A6A
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with NGT | | Descriptor: | 1,2-ETHANEDIOL, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5AC4
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A69
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-PUGNAc | | Descriptor: | N-ACETYL-BETA-D-GLUCOSAMINIDASE, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
