4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6DNO
 
 | | Crystal structure of Protein Phosphatase 1 (PP1) bound to the muscle glycogen-targeting subunit (Gm) | | Descriptor: | Microcystin-LR, Protein phosphatase 1 regulatory subunit 3A, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme.
Sci Adv, 4, 2018
|
|
4Y14
 
 | | Structure of protein tyrosine phosphatase 1B complexed with inhibitor (PTP1B:CPT157633) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-bromo-4-[difluoro(phosphono)methyl]-N-methyl-Nalpha-(methylsulfonyl)-L-phenylalaninamide, CHLORIDE ION, ... | | Authors: | Choy, M.S, Connors, C, Page, R, Peti, W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PTP1B inhibition suggests a therapeutic strategy for Rett syndrome.
J.Clin.Invest., 125, 2015
|
|
5JPF
 
 | | Serine/Threonine phosphatase Z1 (Candida albicans) binds to inhibitor microcystin-LR | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Microcystin-LR, ... | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39929724 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
5JPE
 
 | | Yeast-specific serine/threonine protein phosphatase (PPZ1) of Candida albicans | | Descriptor: | CITRATE ANION, GLYCEROL, Serine/threonine-protein phosphatase | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
8DWL
 
 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8DWK
 
 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
4PHL
 
 | | TbrPDEB1-inhibitor complex | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, Class 1 phosphodiesterase PDEB1, ETHANOL, ... | | Authors: | Choy, M.S, Bland, N, Peti, W, Page, R. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TbrPDEB1-inhibitor complex
To Be Published
|
|
6HKW
 
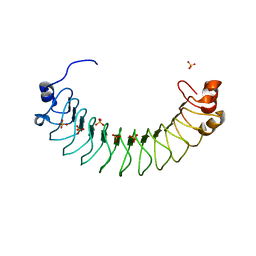 | | Crystal structure of human SDS22 | | Descriptor: | Protein phosphatase 1 regulatory subunit 7, SULFATE ION | | Authors: | Heroes, E, Choy, M.S, Page, R, Peti, W, Ulens, C, Van Meervelt, L, Nys, M, Bollen, M. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Guided Exploration of SDS22 Interactions with Protein Phosphatase PP1 and the Splicing Factor BCLAF1.
Structure, 27, 2019
|
|
8F3R
 
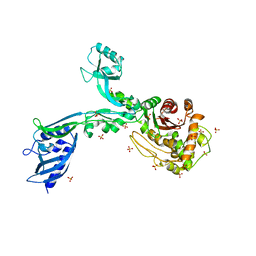 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3G
 
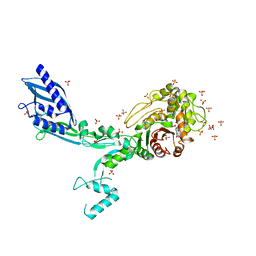 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant in the penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3F
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Hunashal, Y, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3N
 
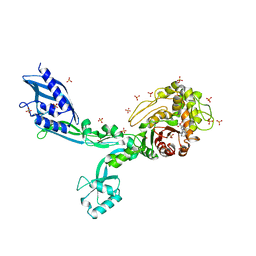 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3P
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3J
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3S
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F67
 
 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | Descriptor: | Pbp5, SULFATE ION | | Authors: | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3M
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3O
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3U
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3I
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3H
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3L
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3T
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
