6CF1
 
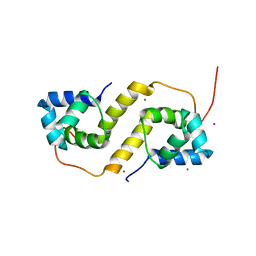 | | Proteus vulgaris HigA antitoxin structure | | 分子名称: | Antitoxin HigA, POTASSIUM ION | | 著者 | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | 登録日 | 2018-02-13 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
4NJN
 
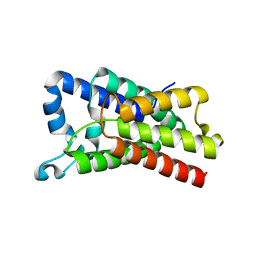 | | Crystal Structure of E.coli GlpG at pH 4.5 | | 分子名称: | Rhomboid protease GlpG | | 著者 | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | 登録日 | 2013-11-11 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
4NJP
 
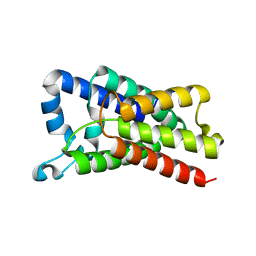 | | Proteolysis inside the membrane is a rate-governed reaction not Driven by substrate affinity | | 分子名称: | Rhomboid protease GlpG | | 著者 | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | 登録日 | 2013-11-11 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
2PP4
 
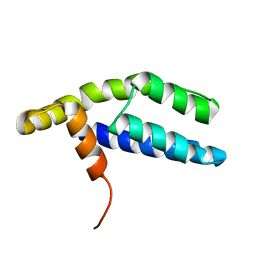 | | Solution Structure of ETO-TAFH refined in explicit solvent | | 分子名称: | Protein ETO | | 著者 | Wei, Y, Liu, S, Lausen, J, Woodrell, C, Cho, S, Biris, N, Kobayashi, N, Yokoyama, S, Werner, M.H. | | 登録日 | 2007-04-27 | | 公開日 | 2007-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription
Nat.Struct.Mol.Biol., 14, 2007
|
|
3MC0
 
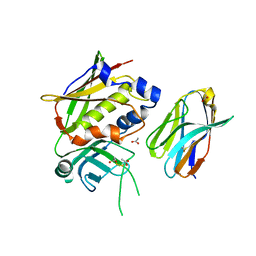 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a Mouse T-cell Receptor beta Chain | | 分子名称: | ACETATE ION, Enterotoxin SEG, variable beta 8.2 mouse T cell receptor | | 著者 | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, E.L. | | 登録日 | 2010-03-26 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
3OWE
 
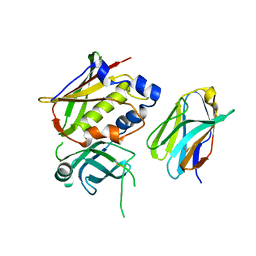 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a High Affinity Mutant Mouse T-cell Receptor Chain | | 分子名称: | Beta-chain, Enterotoxin SEG | | 著者 | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, M.L. | | 登録日 | 2010-09-17 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
1YWO
 
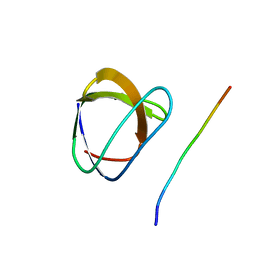 | | Phospholipase Cgamma1 SH3 in complex with a SLP-76 motif | | 分子名称: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1, Lymphocyte cytosolic protein 2 | | 著者 | Deng, L, Velikovsky, C.A, Swaminathan, C.P, Cho, S, Mariuzza, R.A. | | 登録日 | 2005-02-18 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis for Recognition of the T Cell Adaptor Protein SLP-76 by the SH3 Domain of Phospholipase Cgamma1
J.Mol.Biol., 352, 2005
|
|
1YWP
 
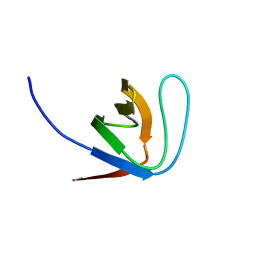 | | Phospholipase Cgamma1 SH3 | | 分子名称: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 | | 著者 | Deng, L, Velikovsky, C.A, Swaminathan, C.P, Cho, S, Mariuzza, R.A. | | 登録日 | 2005-02-18 | | 公開日 | 2005-08-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for Recognition of the T Cell Adaptor Protein SLP-76 by the SH3 Domain of Phospholipase Cgamma1
J.Mol.Biol., 352, 2005
|
|
6BUM
 
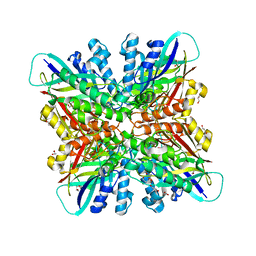 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica | | 分子名称: | 1,3-PROPANDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Shi, K, Cho, S, Seffernick, J.L, Bera, A, Wackett, L.P, Aihara, H. | | 登録日 | 2017-12-11 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
5XBK
 
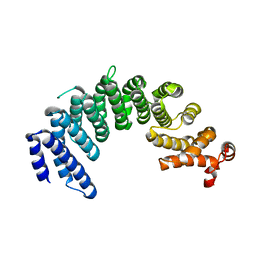 | | Crystal structure of human Importin4 | | 分子名称: | Importin-4, histone H3 | | 著者 | Song, J.J, Yoon, J. | | 登録日 | 2017-03-20 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.223 Å) | | 主引用文献 | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
5XAH
 
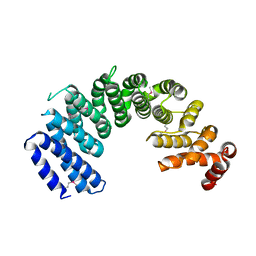 | | Crystal structure of human Importin4 | | 分子名称: | Importin-4 | | 著者 | Song, J.J, Yoon, J. | | 登録日 | 2017-03-13 | | 公開日 | 2018-02-14 | | 最終更新日 | 2018-04-11 | | 実験手法 | X-RAY DIFFRACTION (3.004 Å) | | 主引用文献 | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
1COJ
 
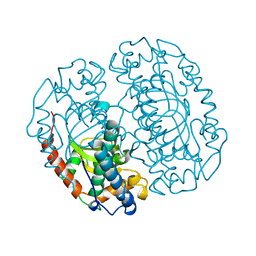 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | 分子名称: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | 著者 | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | 登録日 | 1999-05-28 | | 公開日 | 1999-06-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
3QID
 
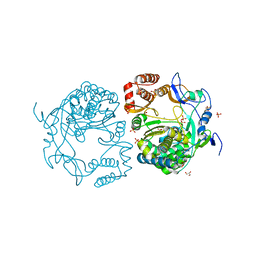 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | 分子名称: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | 著者 | Kim, K.H, Intekhab, A, Lee, J.H. | | 登録日 | 2011-01-27 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
1Z57
 
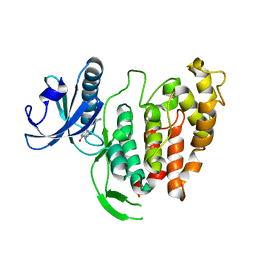 | | Crystal structure of human CLK1 in complex with 10Z-Hymenialdisine | | 分子名称: | DEBROMOHYMENIALDISINE, Dual specificity protein kinase CLK1 | | 著者 | Debreczeni, J, Das, S, Knapp, S, Bullock, A, Guo, K, Amos, A, Fedorov, O, Edwards, A, Sundstrom, M, von Delft, F, Niesen, F.H, Ball, L, Sobott, F, Arrowsmith, C, Structural Genomics Consortium (SGC) | | 登録日 | 2005-03-17 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation.
Structure, 17, 2009
|
|
4FJS
 
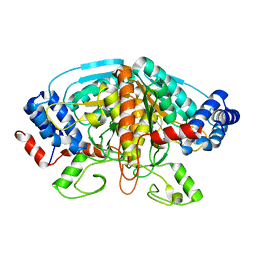 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | 分子名称: | Ureidoglycolate dehydrogenase | | 著者 | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | 登録日 | 2012-06-12 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
4FJU
 
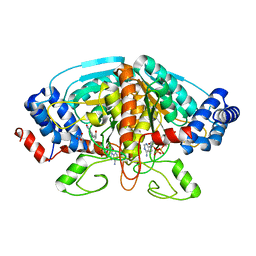 | |
4H8A
 
 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | 著者 | Rhee, S, Shin, I, Kim, M. | | 登録日 | 2012-09-22 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
6CHV
 
 | | Proteus vulgaris HigA antitoxin bound to DNA | | 分子名称: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | 著者 | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6CN8
 
 | |
6CWJ
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | 分子名称: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2018-03-30 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.253 Å) | | 主引用文献 | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6DHJ
 
 | |
1I69
 
 | |
1I6A
 
 | |
6PBA
 
 | | Structure of ClpC1-NTD | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpC1 | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-06-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBQ
 
 | | Structure of ClpC1-NTD | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp protease ATP-binding subunit ClpC1, PHOSPHATE ION | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-06-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
