2R69
 
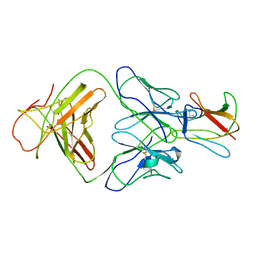 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2R6P
 
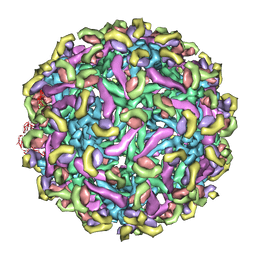 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3IYA
 
 | | Association of the pr peptides with dengue virus blocks membrane fusion at acidic pH | | Descriptor: | Envelope protein, prM protein | | Authors: | Yu, I, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2009-06-01 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion.
J.Virol., 83, 2009
|
|
3IY0
 
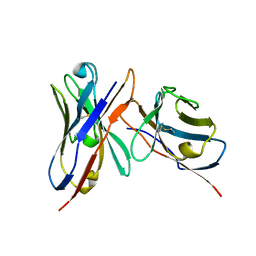 | | Variable domains of the x-ray structure of Fab 14 fitted into the cryoEM reconstruction of the virus-Fab 14 complex | | Descriptor: | Fab 14, heavy domain, light domain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-07 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IXY
 
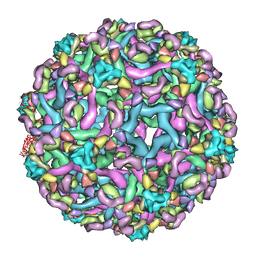 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
3IY7
 
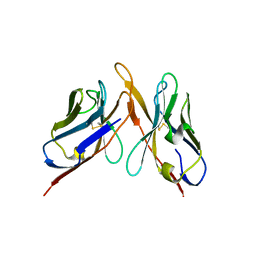 | | Variable domains of the computer generated model (WAM) of Fab F fitted into the cryoEM reconstruction of the virus-Fab F complex | | Descriptor: | fragment from neutralizing antibody F (heavy chain), fragment from neutralizing antibody F (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY2
 
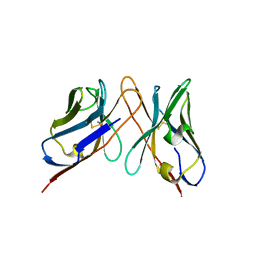 | | Variable domains of the computer generated model (WAM) of Fab 6 fitted into the cryoEM reconstruction of the virus-Fab 6 complex | | Descriptor: | Antibody 6, heavy chain, light chain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IXX
 
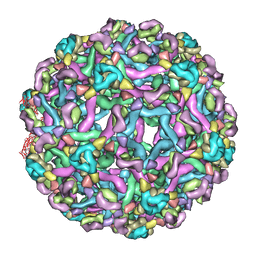 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
3J0B
 
 | | cryo-EM reconstruction of West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, W, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Membrane curvature in flaviviruses.
J.Struct.Biol., 183, 2013
|
|
3IY4
 
 | | Variable domains of the computer generated model (WAM) of Fab 15 fitted into the cryoEM reconstruction of the virus-Fab 15 complex | | Descriptor: | fragment of neutralizing antibody 15 (heavy chain), fragment of neutralizing antibody 15 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IYP
 
 | | The Interaction of Decay-accelerating Factor with Echovirus 7 | | Descriptor: | Capsid protein, Complement decay-accelerating factor, LAURIC ACID, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Harris, K.G, Cifuente, J.O, Bowman, V.D, Chipman, P.R, Lin, F, Medof, D.E, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-04-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Interaction of decay-accelerating factor with echovirus 7.
J.Virol., 84, 2010
|
|
3IY6
 
 | | Variable domains of the computer generated model (WAM) of Fab E fitted into the cryoEM reconstruction of the virus-Fab E complex | | Descriptor: | fragment from neutralizing antibody E (heavy chain), fragment from neutralizing antibody E (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY5
 
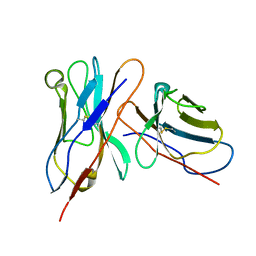 | | Variable domains of the mouse Fab (1AIF) fitted into the cryoEM reconstruction of the virus-Fab 16 complex | | Descriptor: | antibody fragment IGG2A (heavy chain), antibody fragment IGG2A (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY3
 
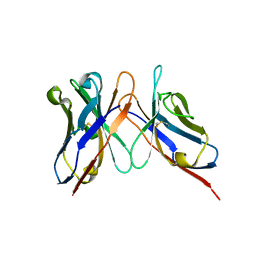 | | Variable domains of the computer generated model (WAM) of Fab 8 fitted into the cryoEM reconstruction of the virus-Fab 8 complex | | Descriptor: | antibody fragment from neutralizing antibody 8 (heavy chain), antibody fragment from neutralizing antibody 8 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1N6G
 
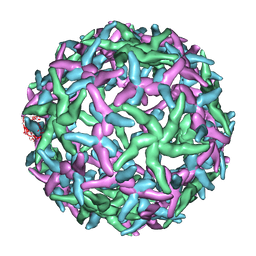 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
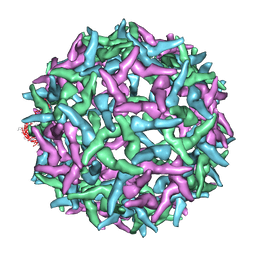 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
1N80
 
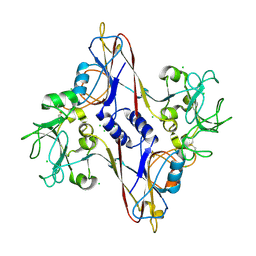 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N7Z
 
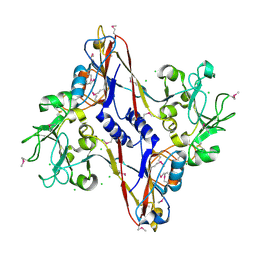 | | Structure and location of gene product 8 in the bacteriophage T4 baseplate | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N8B
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | BROMIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1NN8
 
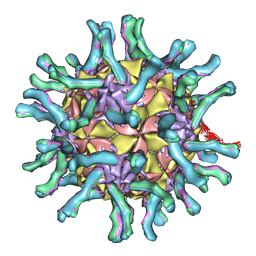 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
1PDF
 
 | | Fitting of gp11 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 baseplate-tail tube complex | | Descriptor: | Baseplate structural protein Gp11 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDI
 
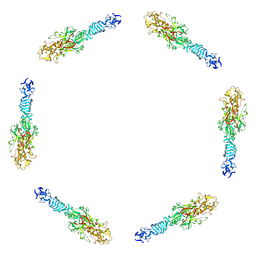 | | Fitting of the C-terminal part of the short tail fibers into the cryo-EM reconstruction of T4 baseplate | | Descriptor: | Short tail fiber protein | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDP
 
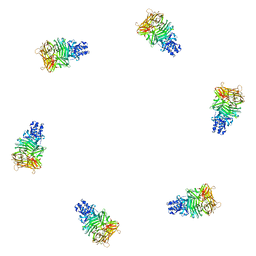 | | Fitting of gp9 structure into the bacteriophage T4 baseplate cryoEM reconstruction | | Descriptor: | Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1KVP
 
 | |
1DNV
 
 | | PARVOVIRUS (DENSOVIRUS) FROM GALLERIA MELLONELLA | | Descriptor: | GALLERIA MELLONELLA DENSOVIRUS CAPSID PROTEIN | | Authors: | Simpson, A.A, Chipmann, P.R, Baker, T.S, Tijssen, P, Rossmann, M.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution.
Structure, 6, 1998
|
|
