7E4K
 
 | |
5XOV
 
 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | Descriptor: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOT
 
 | | Crystal structure of pHLA-B35 in complex with TU55 T cell receptor | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOS
 
 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
6AH3
 
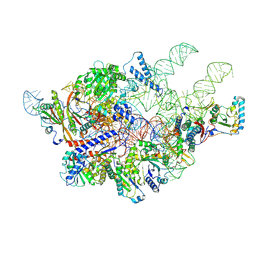 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6AGB
 
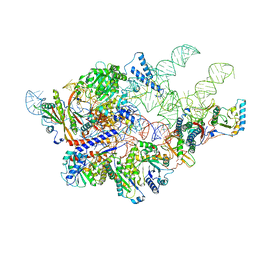 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6CEV
 
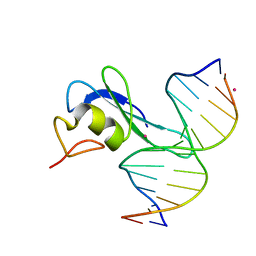 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CEU
 
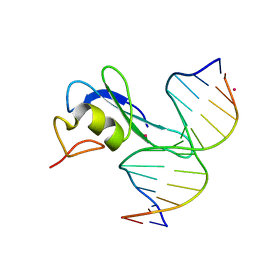 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CC8
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION, methylated CpG DNA | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CCG
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA, Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6IEX
 
 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
6J2A
 
 | | The structure of HLA-A*3003/NP44 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, NP44 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1W
 
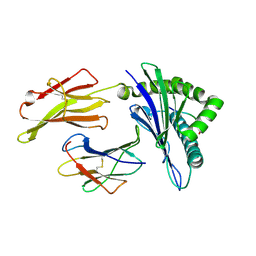 | | The structure of HLA-A*3001/RT313 | | Descriptor: | ALA-ILE-PHE-GLN-SER-SER-MET-THR-LYS, Beta-2-microglobulin, HLA-A*3001 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J29
 
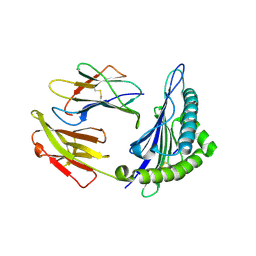 | | The structure of HLA-A*3003/MTB | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, MTB | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1V
 
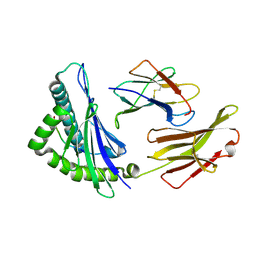 | | The structure of HLA-A*3003/RT313 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, RT313 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8YLE
 
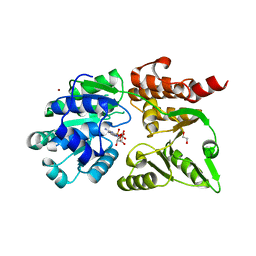 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
8HRI
 
 | |
8HRJ
 
 | |
