9M6H
 
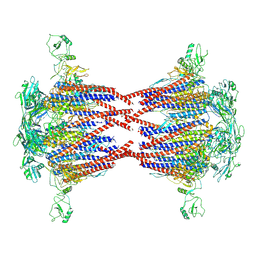 | | structure of FliD-FliC at a 10:10 stoichiometry | | Descriptor: | Flagellar hook-associated protein 2, Flagellin | | Authors: | Chen, L.X, Chen, X.Q, Zhang, F.F, Jiang, W.X, Xing, Q. | | Deposit date: | 2025-03-07 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the initiation of bacterial flagellar filament elongation
To Be Published
|
|
8Z5A
 
 | | Crystal Structure of Nur77 LBD in complex with NLM1 | | Descriptor: | 5-[[4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidin-2-yl]amino]-~{N}-(3-morpholin-4-ylpropyl)-1~{H}-indole-2-carboxamide, GLYCEROL, Nuclear receptor subfamily 4immunitygroup A member 1 | | Authors: | Hong, W.B, Lin, T.W, Chen, X.Q. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Crystal Structure of Nur77 LBD in complex with NLM1
To Be Published
|
|
9IJK
 
 | |
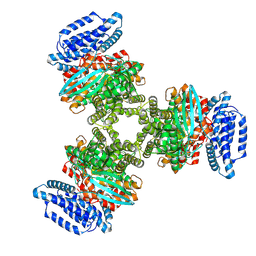
8JZL
 
 | |
8K0A
 
 | |
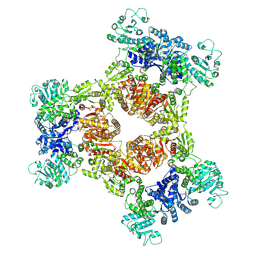
8JZO
 
 | |
8X7F
 
 | | CryoEM structure of the trifunctional NAD biosynthesis/regulator protein NadR in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Trifunctional NAD biosynthesis/regulator protein NadR | | Authors: | Jiang, W.X, Dong, X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-11-24 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | CryoEM structure of the trifunctional NAD biosynthesis/regulator protein NadR in complex with NAD
To Be Published
|
|
8ZPI
 
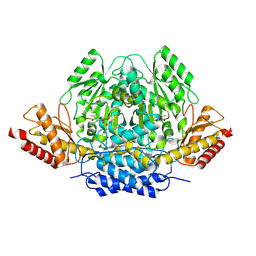 | |
8ZTW
 
 | |
8ZU0
 
 | | CryoEM structure of a tRNA uridine 5-carboxymethylaminomethyl modification enzyme GidA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG | | Authors: | Jiang, W.X, Cheng, X.Q, Dong, X, Ma, L.X, Xing, Q. | | Deposit date: | 2024-06-07 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | CryoEM structure of a tRNA uridine 5-carboxymethylaminomethyl modification enzyme GidA
To Be Published
|
|
8ZU1
 
 | |
7T79
 
 | |
7T78
 
 | |
7YGI
 
 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
7JOU
 
 | |
7JOV
 
 | |
8WUY
 
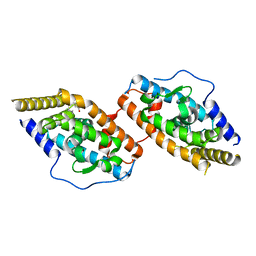 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
8K03
 
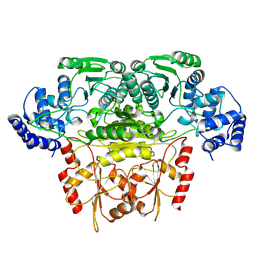 | |
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural and catalytic insights into MhpB: A dioxygenase enzyme for degrading catecholic pollutants.
J Hazard Mater, 488, 2025
|
|
