1WTO
 
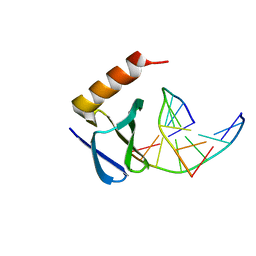 | | Hyperthermophile chromosomal protein SAC7D double mutant V26F/M29F in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTX
 
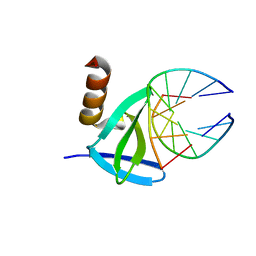 | | Hyperthermophile chromosomal protein SAC7D single mutant V26A in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTP
 
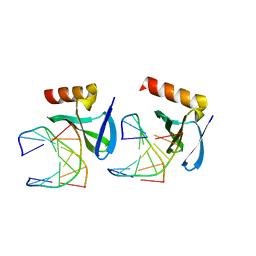 | | Hyperthermophile chromosomal protein SAC7D single mutant M29F in complex with DNA GCGA(UBr)CGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*(BRU)P*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTR
 
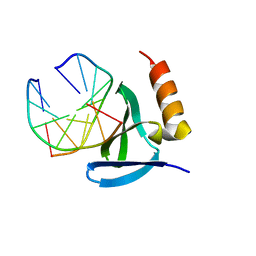 | | Hyperthermophile chromosomal protein SAC7D single mutant M29A in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTW
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant V26A in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTV
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant M29A in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTQ
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant M29F in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1XYI
 
 | | Hyperthermophile chromosomal protein Sac7d double mutant Val26Ala/Met29Ala in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-10 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
Nucleic Acids Res., 33, 2005
|
|
2CJR
 
 | |
4I2T
 
 | |
4I2U
 
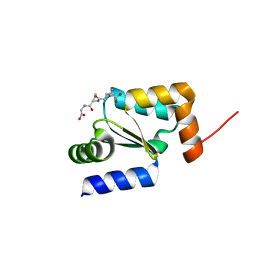 | | Crystal structure of the reduced glutaredoxin from Chlorella sorokiniana T-89 in complex with glutathione | | Descriptor: | GLUTATHIONE, glutaredoxin | | Authors: | Nien, C.-Y, Cheng, C.-Y, Shaw, J.-F, Chen, Y.-T, Liu, J.-H. | | Deposit date: | 2012-11-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional analysis of glutaredoxin from Chlorella sorokiniana T-89
To be Published
|
|
1MR6
 
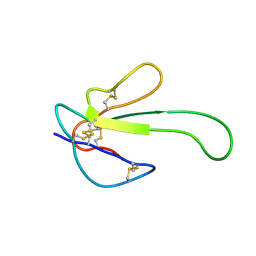 | | Solution Structure of gamma-Bungarotoxin:Implication for the role of the Residues Adjacent to RGD in Integrin Binding | | Descriptor: | neurotoxin | | Authors: | Chuang, W.-J, Shiu, J.-H, Chen, C.-Y, Chen, Y.-C, Chang, L.-S. | | Deposit date: | 2002-09-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma-bungarotoxin: The functional significance of amino acid residues flanking the RGD motif in integrin binding
Proteins, 57, 2004
|
|
1XG7
 
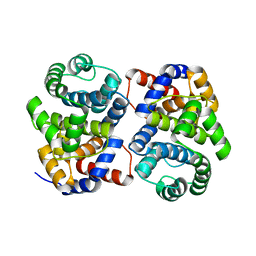 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1R0M
 
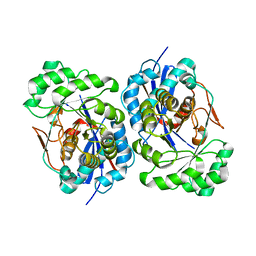 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
5FNN
 
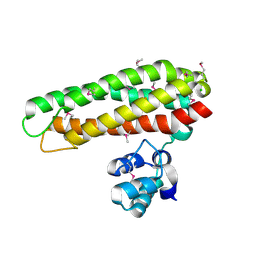 | | Iron and Selenomethionine containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, FE (II) ION, FE (III) ION, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNY
 
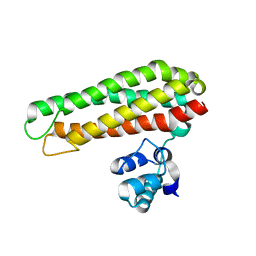 | | Low solvent content crystal form of Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | FE (III) ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, NONAETHYLENE GLYCOL, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNP
 
 | | High resolution Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, OXYGEN ATOM, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
1XPY
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XS2
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
2PJI
 
 | |
1FO6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE | | Descriptor: | N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE, XENON | | Authors: | Wang, W.-C, Hsu, W.-H, Chien, F.-T, Chen, C.-Y. | | Deposit date: | 2000-08-25 | | Release date: | 2001-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft.
J.Mol.Biol., 306, 2001
|
|
3UDA
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI24) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD9
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI23) complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD7
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI21) complexes | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD8
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI22) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
