2JM2
 
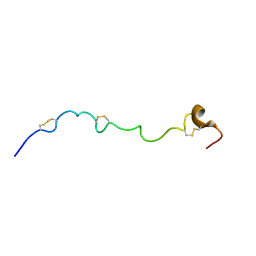 | | Structure of the N-terminal subdomain of insulin-like growth factor (IGF) binding protein-6 and its interactions with IGFs | | Descriptor: | Insulin-like growth factor-binding protein 6 | | Authors: | Chandrashekaran, I.R, Yao, S, Wang, C.C, Bansal, P.S, Alewood, P.F, Forbes, B.E, Wallace, J.C, Bach, L.A, Norton, R.S. | | Deposit date: | 2006-09-18 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Subdomain of Insulin-like Growth Factor (IGF) Binding Protein 6. Structure and Interaction with IGFs
Biochemistry, 46, 2007
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
7N3K
 
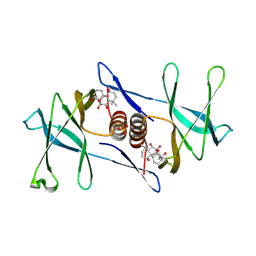 | | Oridonin-bound SARS-CoV-2 Nsp9 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein.
J.Biol.Chem., 297, 2021
|
|
4R1B
 
 | |
4R1C
 
 | |
4R19
 
 | |
4R1A
 
 | |
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DO7
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
8D12
 
 | |
8CZM
 
 | |
8DG0
 
 | |
8CXD
 
 | |
8DG1
 
 | |
8CZN
 
 | |
8D11
 
 | |
8CXE
 
 | |
8D10
 
 | |
8DG2
 
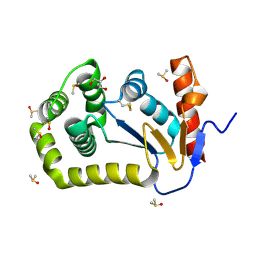 | | Crystal Structure of EcDsbA in a complex with DMSO | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Whitehouse, R.L, Ilyichova, O.V, Taylor, A.J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment screening libraries for the identification of protein hot spots and their minimal binding pharmacophores.
Rsc Med Chem, 14, 2023
|
|
8DQU
 
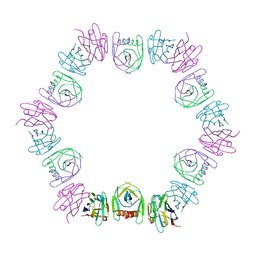 | | Nanobody bound SARS-CoV-2 Nsp9 | | Descriptor: | Nanobody, Non-structural protein 9 | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45003176 Å) | | Cite: | Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9.
Plos One, 18, 2023
|
|
