4S2S
 
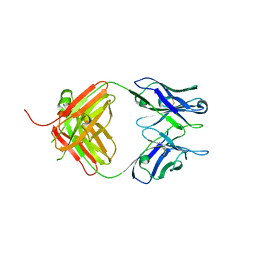 | | Crystal Structure of Fab fragment of monoclonal antibody RoAb13 | | Descriptor: | RoAb13 Fab Heavy chain, RoAb13 Fab Light chain | | Authors: | Chain, B, Arnold, J, Akthar, S, Noursadeghi, M, Lapp, T, Ji, C, Naider, D, Zhang, Y, Govada, L, Saridakis, E, Chayen, N.E. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody.
Plos One, 10, 2015
|
|
1Y7J
 
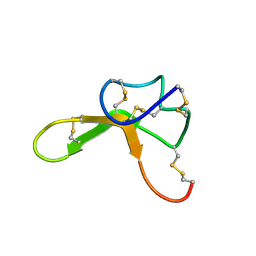 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
1Y7K
 
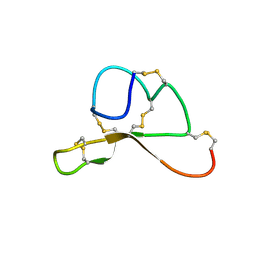 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
1MR0
 
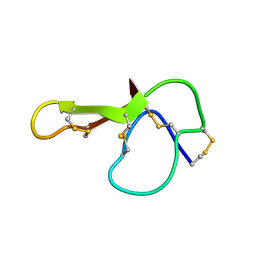 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
6BBM
 
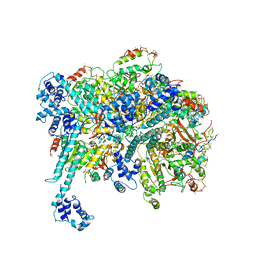 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | Authors: | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | Deposit date: | 2017-10-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
6SHG
 
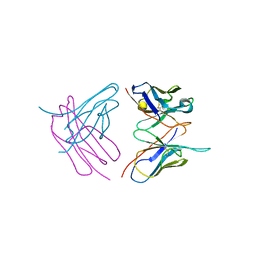 | | Diffraction data for RoAb13 crystal co-crystallised with PIYDIN and its RoAb13 structure | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chain, B, Morgan, M, Kassen, S.C, Chayen, N.E. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.35050821 Å) | | Cite: | X-ray crystallography based model of the antibody RoAb13 bound to peptidomimetics of the HIV receptor chemokine receptor CCR5
To Be Published
|
|
8J1V
 
 | |
8J1T
 
 | |
6PNK
 
 | |
6XEZ
 
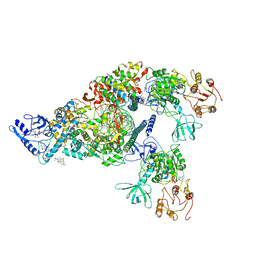 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
6P45
 
 | |
7NJZ
 
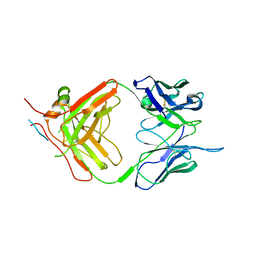 | | X-ray crystallography study of RoAb13 which binds to PIYDIN, a part of the CCR5 N terminal domain | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Helliwell, J.R, Chayen, N, Saridakis, E, Govada, L. | | Deposit date: | 2021-02-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
7NW3
 
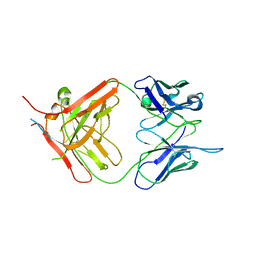 | | X-ray crystallographic study of PIYDIN, which contains the truncation determinants of binding PI and N, bound to RoAb13, a CCR5 antibody | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chayen, N.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.200011 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
1V1O
 
 | |
1V1P
 
 | |
4ZSW
 
 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4ZSY
 
 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
