5IR0
 
 | | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant | | Descriptor: | CITRIC ACID, Uncharacterized protein ORF19 | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (3.297 Å) | | Cite: | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant
To Be Published
|
|
5KP2
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
5TVL
 
 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
4LES
 
 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
5FDA
 
 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
3KBO
 
 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
3KB8
 
 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3L44
 
 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
6W34
 
 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus
To Be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
3RU6
 
 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3R8Y
 
 | | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, CALCIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase
TO BE PUBLISHED
|
|
6VJ2
 
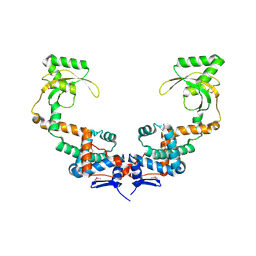 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
3RE3
 
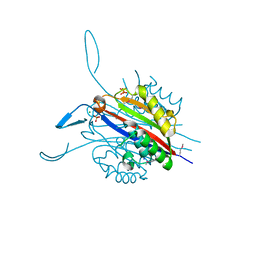 | | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-02 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis
To be Published
|
|
6VJ4
 
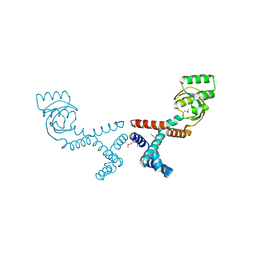 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6OZ7
 
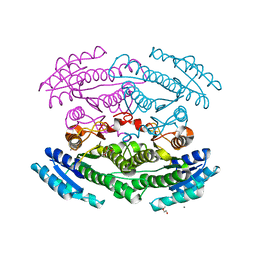 | | Putative oxidoreductase from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-15 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
3R3T
 
 | | Crystal Structure of 30S Ribosomal Protein S from Bacillus anthracis | | Descriptor: | 30S ribosomal protein S6, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of 30S Ribosomal Protein S from Bacillus anthracis
To be Published
|
|
3RJU
 
 | | Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate | | Descriptor: | Beta-lactamase/D-alanine Carboxypeptidase, CITRIC ACID, GLYCEROL | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate
To be Published
|
|
3RPE
 
 | | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Modulator of drug activity B | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
6PFN
 
 | | Succinyl-CoA synthase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COENZYME A, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Succinyl-CoA synthase from Francisella tularensis
to be published
|
|
4XOX
 
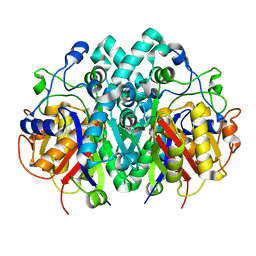 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
3R2I
 
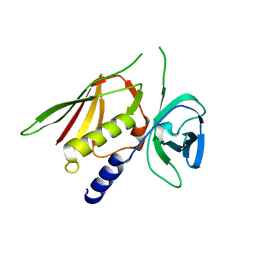 | | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Exotoxin 5 | | Authors: | Filippova, E.V, Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3RUY
 
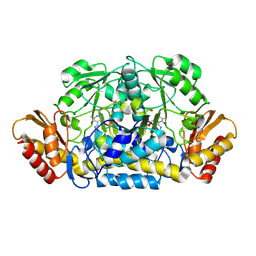 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | Descriptor: | Ornithine aminotransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
7SGW
 
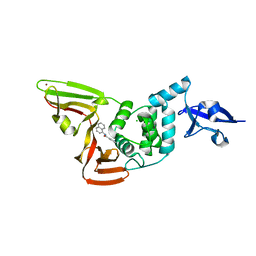 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
