1F6D
 
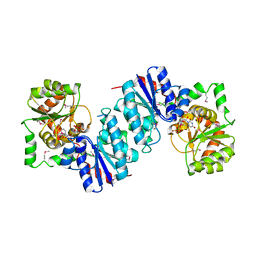 | | THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI. | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE, ... | | Authors: | Campbell, R.E, Mosimann, S.C, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases.
Biochemistry, 39, 2000
|
|
1DLI
 
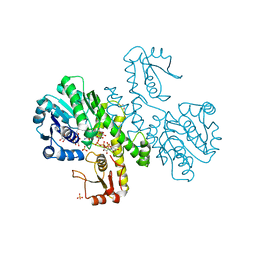 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1DLJ
 
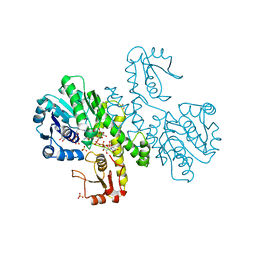 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
2ORU
 
 | |
7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
7E9Y
 
 | | Crystal structure of eLACCO1 | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CALCIUM ION, Lactate-binding periplasmic protein TTHA0766,Lactate-binding periplasmic protein TTHA0766 | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J, Nasu, Y. | | Deposit date: | 2021-03-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A genetically encoded fluorescent biosensor for extracellular L-lactate.
Nat Commun, 12, 2021
|
|
8ZEX
 
 | | Biosensor HaloKbp1a | | Descriptor: | HaloKbp1a, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E. | | Deposit date: | 2024-05-07 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | High-Performance Chemigenetic Potassium Ion Indicator.
J.Am.Chem.Soc., 2024
|
|
3IP2
 
 | | Crystal structure of red fluorescent protein Neptune at pH 7.0 | | Descriptor: | Neptune red fluorescent protein | | Authors: | Lin, M.Z, McKeown, M.R, Ng, H.L, Aguilera, T.A, Shaner, N.C, Ma, W, Adams, S.R, Campbell, R.E, Alber, T, Tsien, R.Y. | | Deposit date: | 2009-08-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals.
Chem.Biol., 16, 2009
|
|
2OTB
 
 | |
2OTE
 
 | | Crystal structure of a monomeric cyan fluorescent protein in the photobleached state | | Descriptor: | ACETATE ION, GFP-like fluorescent chromoprotein cFP484 | | Authors: | Henderson, J.N, Ai, H, Campbell, R.E, Remington, S.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for reversible photobleaching of a green fluorescent protein homologue.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HQK
 
 | | Crystal structure of a monomeric cyan fluorescent protein derived from Clavularia | | Descriptor: | ACETATE ION, CHLORIDE ION, Cyan fluorescent chromoprotein, ... | | Authors: | Henderson, J.N, Campbell, R.E, Ai, H, Remington, S.J. | | Deposit date: | 2006-07-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging.
Biochem.J., 400, 2006
|
|
1HUY
 
 | | CRYSTAL STRUCTURE OF CITRINE, AN IMPROVED YELLOW VARIANT OF GREEN FLUORESCENT PROTEIN | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Griesbeck, O, Baird, G.S, Campbell, R.E, Zacharias, D.A, Tsien, R.Y. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications
J.Biol.Chem., 276, 2001
|
|
7YV5
 
 | |
7YV3
 
 | |
6LNP
 
 | | Crystal structure of citrate Biosensor | | Descriptor: | CITRIC ACID, Fusion protein of Green fluorescent protein and Sensor histidine kinase CitA | | Authors: | Wen, Y, Campbell, R. | | Deposit date: | 2019-12-31 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | High-Performance Intensiometric Direct- and Inverse-Response Genetically Encoded Biosensors for Citrate.
Acs Cent.Sci., 6, 2020
|
|
7Z7P
 
 | |
7Z7Q
 
 | |
7Z7O
 
 | |
7DMX
 
 | |
7DNA
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
6DEJ
 
 | |
5UKG
 
 | |
