1FR8
 
 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH URIDINE DIPHOSPHOGALACTOSE | | Descriptor: | BETA 1,4 GALACTOSYLTRANSFERASE, GALACTOSE-URIDINE-5'-DIPHOSPHATE | | Authors: | Gastinel, L.N, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-09-20 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
1G9E
 
 | | SOLUTION STRUCTURE AND RELAXATION MEASUREMENTS OF AN ANTIGEN-FREE HEAVY CHAIN VARIABLE DOMAIN (VHH) FROM LLAMA | | Descriptor: | H14 | | Authors: | Renisio, J.-G, Perez, J, Czisch, M, Guenneugues, M, Bornet, O, Frenken, L, Cambillau, C, Darbon, H. | | Deposit date: | 2000-11-23 | | Release date: | 2002-10-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of an antigen-free heavy chain variable
domain (VHH) from Llama
Proteins, 47, 2002
|
|
1FWY
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-25 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
1GJQ
 
 | | Pseudomonas aeruginosa cd1 nitrite reductase reduced cyanide complex | | Descriptor: | CYANIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanide Binding to Cd(1) Nitrite Reductase from Pseudomonas Aeruginosa: Role of the Active-Site His369 in Ligand Stabilization.
Biochem.Biophys.Res.Commun., 291, 2002
|
|
2OXE
 
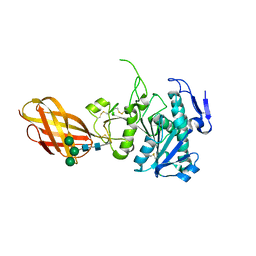 | | Structure of the Human Pancreatic Lipase-related Protein 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic lipase-related protein 2, ... | | Authors: | Walker, J.R, Davis, T, Seitova, A, Finerty Jr, P.J, Butler-Cole, C, Kozieradzki, I, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
7SQX
 
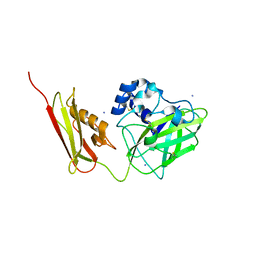 | | Crystal Structure of Pseudomonas aeruginosa lytic polysaccharide monooxygenase CbpD | | Descriptor: | AMMONIUM ION, Chitin-binding protein CbpD | | Authors: | Dade, C, Douzi, B, Ball, G, Voulhoux, R, Forest, K.T. | | Deposit date: | 2021-11-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of CbpD clarifies substrate-specificity motifs in chitin-active lytic polysaccharide monooxygenases.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3GKO
 
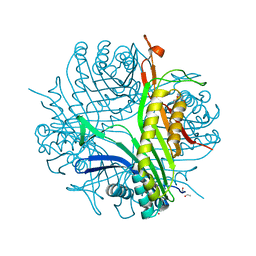 | | Crystal structure of urate oxydase using surfactant Poloxamer 188 as a New Crystallizing Agent | | Descriptor: | 8-AZAXANTHINE, POTASSIUM ION, Uricase | | Authors: | Delfosse, V, Giffard, M, Sciara, G, Bonnete, F, Mayer, C. | | Deposit date: | 2009-03-11 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Surfactant Poloxamer 188 as a New Crystallizing Agent for Urate Oxidase
Cryst.Growth Des., 9, 2009
|
|
5FWO
 
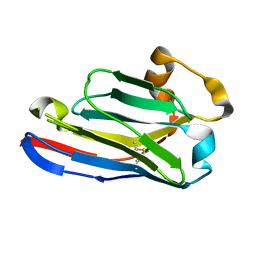 | | Llama nanobody PorM_130 | | Descriptor: | NANOBODY | | Authors: | Leone, P, Roussel, A. | | Deposit date: | 2016-02-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4MKQ
 
 | |
4MJT
 
 | |
4MKO
 
 | |
4ART
 
 | |
4ATS
 
 | |
1ETH
 
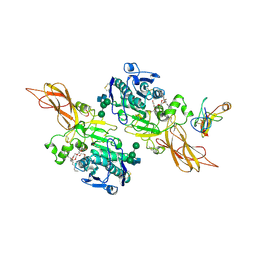 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
5LZ0
 
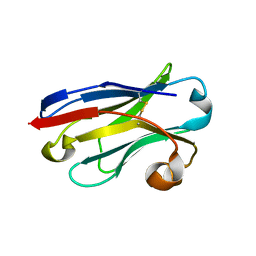 | | Llama nanobody PorM_01 | | Descriptor: | PorM nanobody | | Authors: | Duhoo, Y, Leone, P, Roussel, A. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2BYO
 
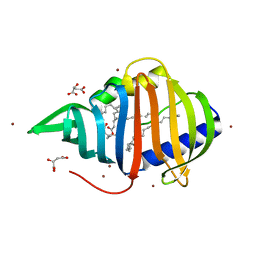 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
2FAV
 
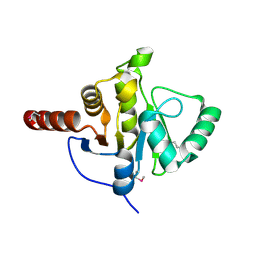 | |
5MP2
 
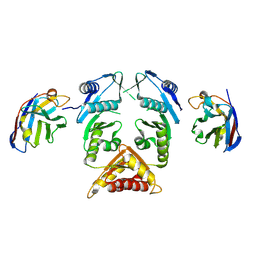 | | XcpQN012 in complex with VHH04 | | Descriptor: | Camelid nanobody VHH04, Type II secretion system protein D | | Authors: | Trinh, T.T.N, Roussel, A, Douzi, B, Voulhoux, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
MBio, 8, 2017
|
|
1OSE
 
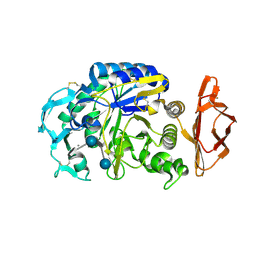 | | Porcine pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gilles, C, Payan, F. | | Deposit date: | 1996-03-20 | | Release date: | 1997-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose.
Eur.J.Biochem., 238, 1996
|
|
1GL0
 
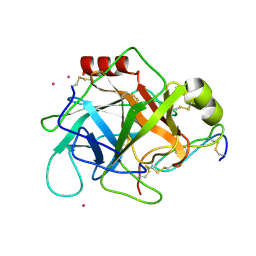 | |
1GL1
 
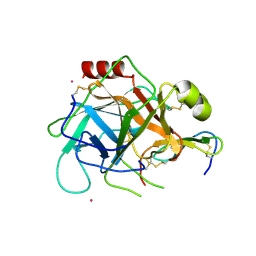 | |
4AX9
 
 | | Human thrombin complexed with Napsagatran, RO0466240 | | Descriptor: | 2-[[(2S)-4-[[(3S)-1-carbamimidoylpiperidin-3-yl]methylamino]-2-(naphthalen-2-ylsulfonylamino)-4-oxidanylidene-butanoyl] -cyclopropyl-amino]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Banner, D.W, D'Arcy, A, Winkler, F.K, Hilpert, K, Spinelli, S, Cambillau, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Potent and Highly Selective Thrombin Inhibitors.
J.Med.Chem., 37, 1994
|
|
