1ELU
 
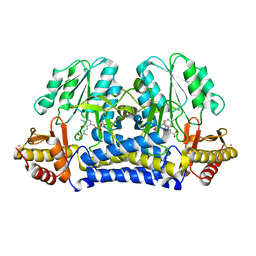 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EUJ
 
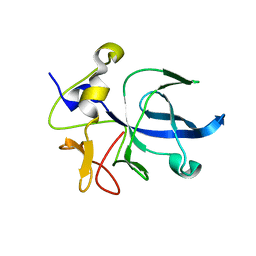 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
1E8L
 
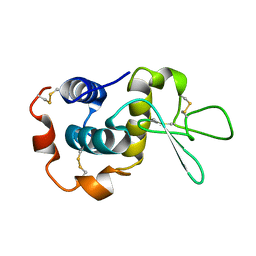 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
1EXL
 
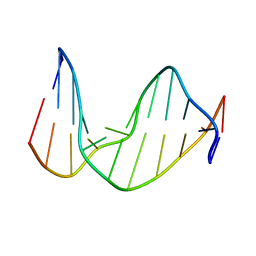 | | STRUCTURE OF AN 11-MER DNA DUPLEX CONTAINING THE CARBOCYCLIC NUCLEOTIDE ANALOG: 2'-DEOXYARISTEROMYCIN | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*(2AR)P*GP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*CP*TP*CP*AP*CP*TP*G)-3') | | Authors: | Smirnov, S, Johnson, F, Marumoto, R, de los Santos, C. | | Deposit date: | 2000-05-03 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an 11-mer DNA duplex containing the carbocyclic nucleotide analog: 2'-deoxyaristeromycin
J.Biomol.Struct.Dyn., 17, 2000
|
|
1ELW
 
 | | Crystal structure of the TPR1 domain of HOP in complex with a HSC70 peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HSC70-PEPTIDE, NICKEL (II) ION, ... | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine
Cell(Cambridge,Mass.), 101, 2000
|
|
1E6O
 
 | | Crystal structure of Fab13B5 against HIV-1 capsid protein p24 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Monaco-Malbet, S, Berthet-Colominas, C, Novelli, A, Battai, N, Piga, N, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-21 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
1ERJ
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL WD40 DOMAIN OF TUP1 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR TUP1 | | Authors: | Sprague, E.R, Redd, M.J, Johnson, A.D, Wolberger, C. | | Deposit date: | 2000-04-06 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast.
EMBO J., 19, 2000
|
|
1ET4
 
 | |
1EC6
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN BOUND TO 20-MER RNA HAIRPIN | | Descriptor: | 20-MER RNA HAIRPIN, RNA-BINDING PROTEIN NOVA-2 | | Authors: | Lewis, H.A, Musunuru, K, Jensen, K.B, Edo, C, Chen, H. | | Deposit date: | 2000-01-25 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome.
Cell(Cambridge,Mass.), 100, 2000
|
|
1EBC
 
 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
1EBL
 
 | | THE 1.8 A CRYSTAL STRUCTURE AND ACTIVE SITE ARCHITECTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE III (FABH) FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL-ACP SYNTHASE III, COENZYME A | | Authors: | Davies, C, Heath, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.
Structure Fold.Des., 8, 2000
|
|
1EGP
 
 | |
1EI5
 
 | | CRYSTAL STRUCTURE OF A D-AMINOPEPTIDASE FROM OCHROBACTRUM ANTHROPI | | Descriptor: | D-AMINOPEPTIDASE | | Authors: | Bompard-Gilles, C, Remaut, H, Villeret, V, Prange, T, Fanuel, L, Joris, J, Frere, J.-M, Van Beeumen, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a D-aminopeptidase from Ochrobactrum anthropi, a new member of the 'penicillin-recognizing enzyme' family.
Structure Fold.Des., 8, 2000
|
|
1E7S
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1EH5
 
 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 COMPLEXED WITH PALMITATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EK9
 
 | | 2.1A X-RAY STRUCTURE OF TOLC: AN INTEGRAL OUTER MEMBRANE PROTEIN AND EFFLUX PUMP COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PROTEIN TOLC | | Authors: | Koronakis, V, Sharff, A.J, Koronakis, E, Luisi, B, Hughes, C. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Nature, 405, 2000
|
|
1ELV
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1S COMPONENT, ... | | Authors: | Gaboriaud, C, Rossi, V, Bally, I, Arlaud, G, Fontecilla-Camps, J.-C. | | Deposit date: | 2000-03-14 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
EMBO J., 19, 2000
|
|
1EB9
 
 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | HYDROXYNITRILE LYASE, P-HYDROXYBENZALDEHYDE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
1EV4
 
 | | RAT GLUTATHIONE S-TRANSFERASE A1-1: MUTANT W21F/F220Y WITH GSO3 BOUND | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, GLUTATHIONE SULFONIC ACID, SULFATE ION | | Authors: | Adman, E.T, Le Trong, I, Stenkamp, R.E, Nieslanik, B.S, Dietze, E.C, Tai, G, Ibarra, C, Atkins, W.M. | | Deposit date: | 2000-04-19 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Localization of the C-terminus of rat glutathione S-transferase A1-1: crystal structure of mutants W21F and W21F/F220Y.
Proteins, 42, 2001
|
|
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
1EF2
 
 | | CRYSTAL STRUCTURE OF MANGANESE-SUBSTITUTED KLEBSIELLA AEROGENES UREASE | | Descriptor: | MANGANESE (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Yamaguchi, K, Cosper, N.J, Stalhandske, C, Scott, R.A, Pearson, M.A, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-02-05 | | Release date: | 2000-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of metal-substituted Klebsiella aerogenes urease.
J.Biol.Inorg.Chem., 4, 1999
|
|
1EFO
 
 | | CRYSTAL STRUCTURE OF AN ADENINE BULGE IN THE RNA CHAIN OF A DNA/RNA HYBRID, D(CTCCTCTTC)/R(GAAGAGAGAG) | | Descriptor: | DNA (5'-D(*CP*TP*CP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3') | | Authors: | Sudarsanakumar, C, Xiong, Y, Sundaralingam, M. | | Deposit date: | 2000-02-09 | | Release date: | 2000-05-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine bulge in the RNA chain of a DNA.RNA hybrid, d(CTCCTCTTC).r(gaagagagag).
J.Mol.Biol., 299, 2000
|
|
1EEQ
 
 | | M4L/Y(27D)D/T94H Mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Raffen, R, Dieckman, L, Boogaard, C, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increasing protein stability by polar surface residues: domain-wide consequences of interactions within a loop.
Biophys.J., 82, 2002
|
|
1ELY
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102C) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
