5HUD
 
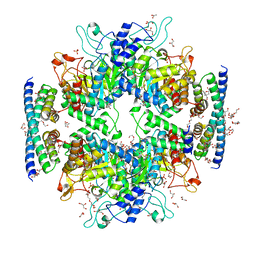 | | Non-covalent complex of and DAHP synthase and chorismate mutase from Corynebacterium glutamicum with bound transition state analog | | Descriptor: | 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUB
 
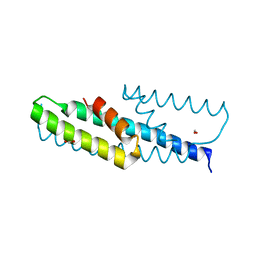 | | High-resolution structure of chorismate mutase from Corynebacterium glutamicum | | Descriptor: | Chorismate mutase, FORMIC ACID | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUE
 
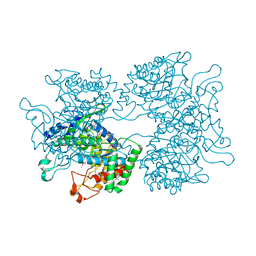 | | DAHP synthase from Corynebacterium glutamicum in complex with tryptophan | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Last modified: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Synthetic alpha-synuclein fibrils replicate in mice causing MSA-like pathology.
Nature, 648, 2025
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP7
 
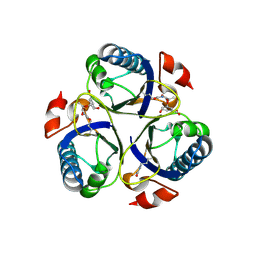 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with chorismate and prephenate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, CHORISMATE MUTASE AROH, PREPHENIC ACID | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP4
 
 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with a transition state analog | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZOP
 
 | | Arg90Cit chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-22 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5HUC
 
 | | DAHP synthase from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, MANGANESE (II) ION, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
2L4F
 
 | |
2L4E
 
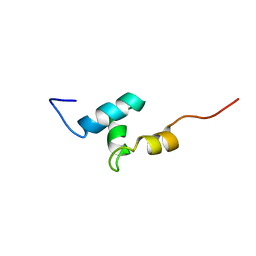 | |
2KWU
 
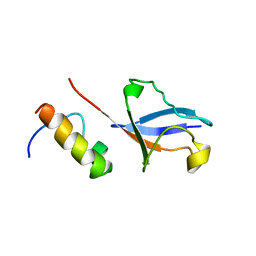 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWV
 
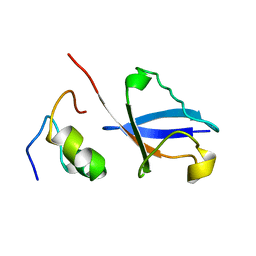 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
5IO9
 
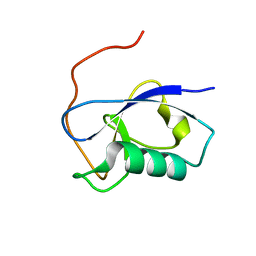 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
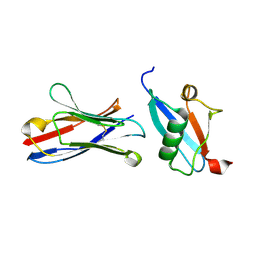 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6I83
 
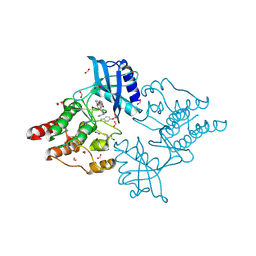 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
6I82
 
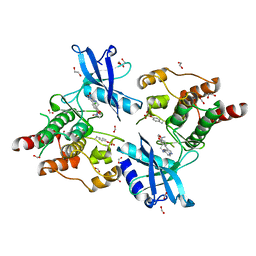 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
9RZF
 
 | | Structure of in-vivo formed alpha-synuclein fibrils purified from a M83+/- mouse brain injected with recombinant 1B fibrils | | Descriptor: | Alpha-synuclein | | Authors: | van den Heuvel, L, Burger, D, Kashyrina, M, de La Seigliere, H, Lewis, A.J, De Nuccio, F, Mohammed, I, Verchere, J, Feuillie, C, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Canron, M, Meissner, W.G, Loquet, A, Bousset, L, Poujol, C, Nilsson, K.P.R, Laferriere, F, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2025-07-15 | | Release date: | 2025-07-30 | | Last modified: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthetic alpha-synuclein fibrils replicate in mice causing MSA-like pathology.
Nature, 648, 2025
|
|
6SS2
 
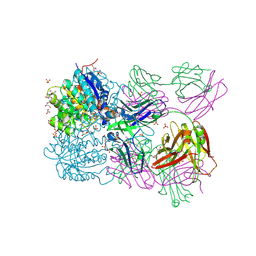 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6TUL
 
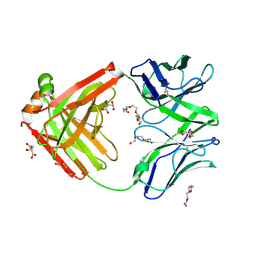 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS5
 
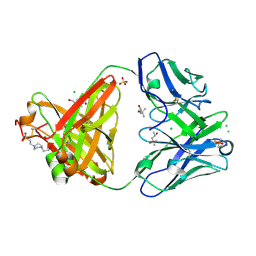 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SRV
 
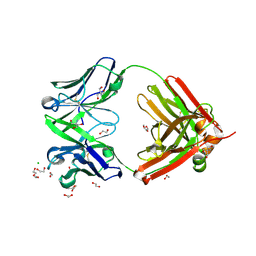 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS0
 
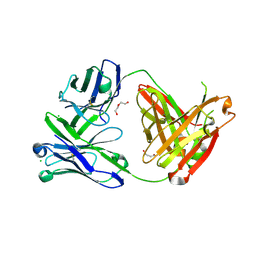 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SRX
 
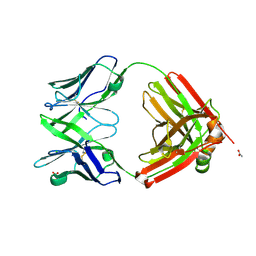 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS6
 
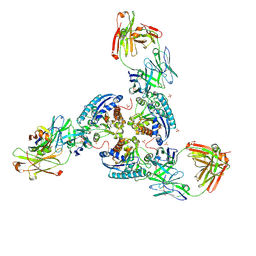 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | Arginase-2, mitochondrial, Fab C0020187 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
