9G15
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O* state, pH 8.8 | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
9G16
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O state obtained by cryotrapping | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
8RSS
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O* state | | Descriptor: | EICOSANE, Microbial rhodopsin, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
8RSR
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) - human GTPase Arf1 (L8K,Q71L) chimera; N state | | Descriptor: | EICOSANE, GUANOSINE-5'-DIPHOSPHATE, Microbial rhodopsin - GTPase chimera,ADP-ribosylation factor 1, ... | | Authors: | Bukhdruker, S, Bratanov, D, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
8RSO
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the ground state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
8RSQ
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) - human GTPase Arf1 (L8K,Q71L) chimera; Ground state | | Descriptor: | EICOSANE, GUANOSINE-5'-DIPHOSPHATE, MIcrobial rhodopsin - GTPase chimera,ADP-ribosylation factor 1, ... | | Authors: | Bukhdruker, S, Bratanov, D, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
8RSP
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the P596 state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
6T0J
 
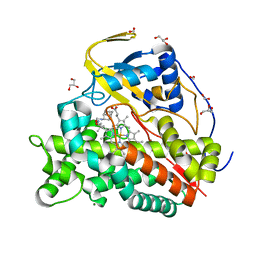 | | Crystal structure of CYP124 in complex with SQ109 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylation of Antitubercular Drug Candidate, SQ109, by Mycobacterial Cytochrome P450.
Int J Mol Sci, 21, 2020
|
|
7ZB9
 
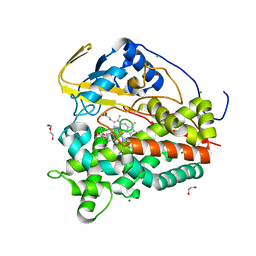 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8YWQ
 
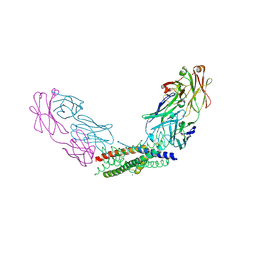 | | Crystal structure of the Fab fragment of the anti-IL-6 antibody I9H in complex with a domain-swapped IL-6 dimer | | Descriptor: | Heavy chain of the Fab fragment of anti-IL-6 antibody I9H, Interleukin-6, Light chain of the Fab fragment of anti-IL-6 antibody I9H, ... | | Authors: | Bukhdruker, S, Yudenko, A, Marin, E, Remeeva, A, Rodin, S, Burtseva, A, Petrov, A, Ischenko, A, Borshchevskiy, V. | | Deposit date: | 2024-03-31 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers.
Structure, 33, 2025
|
|
8YWR
 
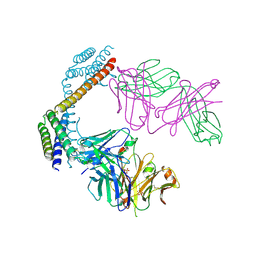 | | Crystal structure of the Fab fragment of the anti-IL-6 antibody 68F2 in complex with a domain-swapped IL-6 dimer | | Descriptor: | Heavy chain of the Fab fragment of anti-IL-6 antibody 68F2, Interleukin-6, Light chain of the Fab fragment of anti-IL-6 antibody 68F2, ... | | Authors: | Bukhdruker, S, Yudenko, A, Borshchevskiy, V. | | Deposit date: | 2024-03-31 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers.
Structure, 33, 2025
|
|
9IN9
 
 | | Structure of the viral channelrhodopsin OLPVR1 at low pH obtained by soaking | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
9IN7
 
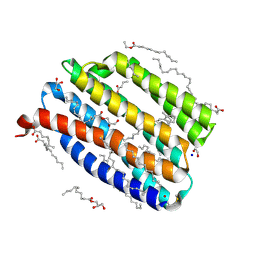 | | True-atomic resolution crystal structure of the closed state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
9IN8
 
 | | True-atomic resolution crystal structure of the open state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
7AV6
 
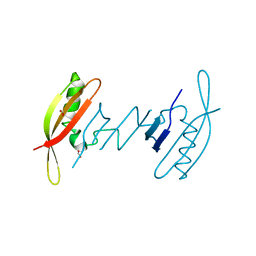 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
6T0L
 
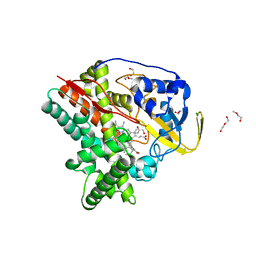 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0K
 
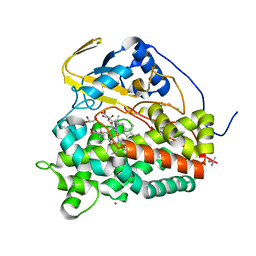 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0F
 
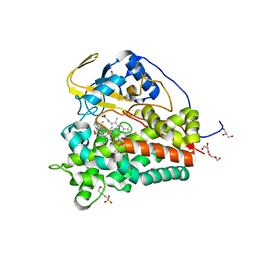 | | Crystal structure of CYP124 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0H
 
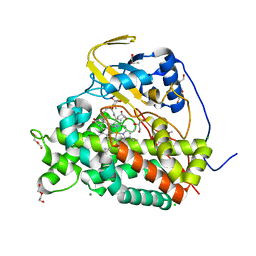 | | Crystal structure of CYP124 in complex with 1-alpha-hydroxy-vitamin D3 | | Descriptor: | 1-alpha-hydroxy-vitamin D3, CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0G
 
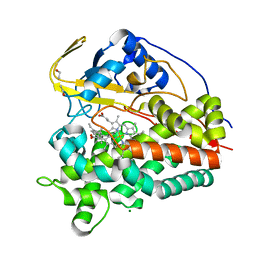 | | Crystal structure of CYP124 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, MAGNESIUM ION, Methyl-branched lipid omega-hydroxylase, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
8AMQ
 
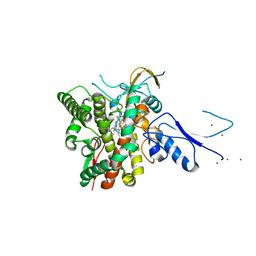 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | Descriptor: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMO
 
 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMP
 
 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
8YWP
 
 | | Crystal structure of the Fab fragment of anti-IL-6 antibody I9H | | Descriptor: | GLYCEROL, Heavy chain of the Fab fragment of anti-IL-6 antibody I9H, Light chain of the Fab fragment of anti-IL-6 antibody I9H, ... | | Authors: | Yudenko, A, Bukhdruker, S, Eliseev, I, Rodin, S, Burtseva, A, Petrov, A, Zlobina, O, Ischenko, A, Borshchevskiy, V. | | Deposit date: | 2024-03-31 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers.
Structure, 33, 2025
|
|
