7K9B
 
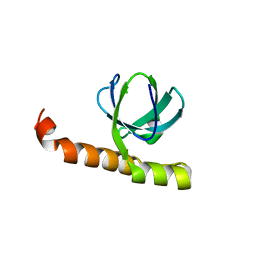 | | Crystal structure of Bacillus halodurans OapB (native) at 1.2 A | | Descriptor: | CHLORIDE ION, OLE-associated protein B | | Authors: | Yang, Y, Breaker, R.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structure of a bacterial OapB protein with its OLE RNA target gives insights into the architecture of the OLE ribonucleoprotein complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KKV
 
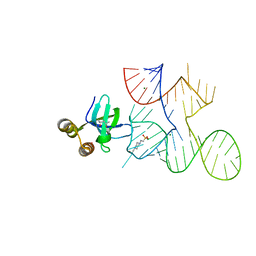 | | Crystal structure of Bacillus halodurans OapB in complex with its OLE RNA target (native, crystal form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Yang, Y, Breaker, R.R. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a bacterial OapB protein with its OLE RNA target gives insights into the architecture of the OLE ribonucleoprotein complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K9E
 
 | |
7K9C
 
 | |
7K9D
 
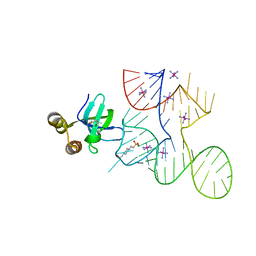 | |
3IRW
 
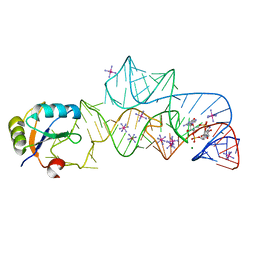 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4RUM
 
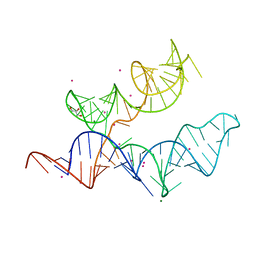 | |
2GDI
 
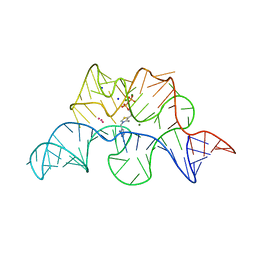 | |
1Y26
 
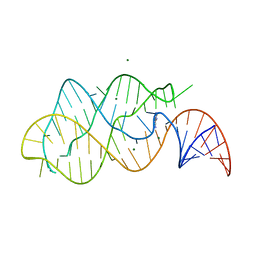 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
1Y27
 
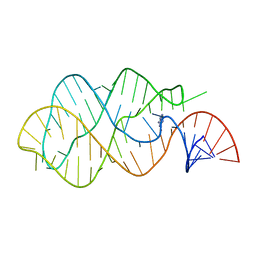 | | G-riboswitch-guanine complex | | Descriptor: | Bacillus subtilis xpt, GUANINE | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
5T83
 
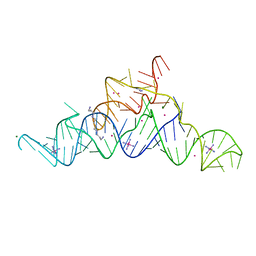 | | Structure of a guanidine-I riboswitch from S. acidophilus | | Descriptor: | GUANIDINE, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Reiss, C.W, Xiong, Y, Strobel, S.A. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Ligand Binding to the Guanidine-I Riboswitch.
Structure, 25, 2017
|
|
