4NUT
 
 | | Crystal structure of the complex between Snu13p and the PEP domain of Rsa1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Ribosome assembly 1 protein, SULFATE ION | | Authors: | Charron, C, Chagot, M.E, Manival, X, Branlant, C, Charpentier, B. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control
J.Cell Biol., 207, 2014
|
|
2AUS
 
 | | Crystal structure of the archaeal box H/ACA sRNP Nop10-Cbf5 complex | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein Nop10, ZINC ION, ... | | Authors: | Charron, C, Manival, X, Charpentier, B, Fourmann, J.-B, Godard, F, Branlant, C. | | Deposit date: | 2005-08-29 | | Release date: | 2006-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity
Nucleic Acids Res., 34, 2006
|
|
1PXW
 
 | | Crystal structure of L7Ae sRNP core protein from Pyrococcus abyssii | | Descriptor: | LSU ribosomal protein L7AE | | Authors: | Charron, C, Manival, X, Charpentier, B, Branlant, C, Aubry, A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2LSV
 
 | | The NMR high resolution structure of yeast Tah1 in complex with the Hsp90 C-terminal tail | | Descriptor: | ATP-dependent molecular chaperone HSP82, TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
2LSU
 
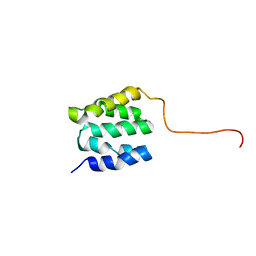 | | The NMR high resolution structure of yeast Tah1 in a free form | | Descriptor: | TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
1GAE
 
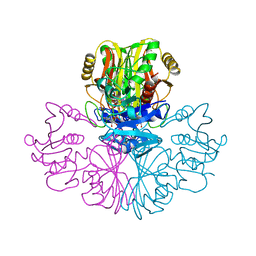 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1GAD
 
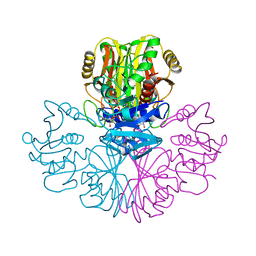 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
2DBV
 
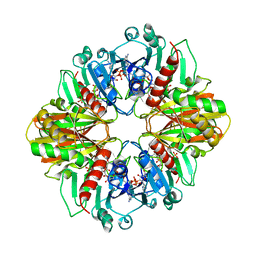 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
6ZBK
 
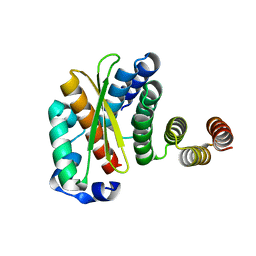 | | Crystal structure of the human complex between RPAP3 and TRBP | | Descriptor: | RISC-loading complex subunit TARBP2, RNA polymerase II-associated protein 3 | | Authors: | Charron, C, Abel, Y, Charpentier, B, Rederstorff, M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Nucleic Acids Res., 50, 2022
|
|
1DBV
 
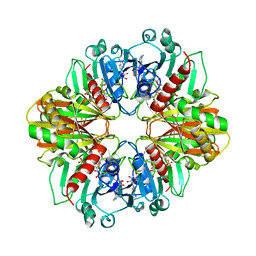 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
3DBV
 
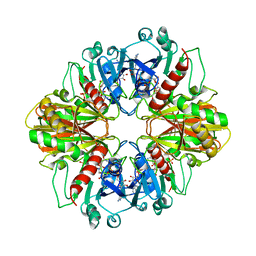 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1GD1
 
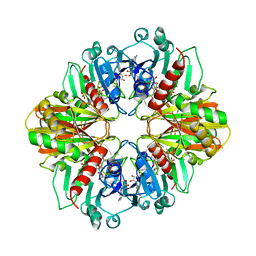 | |
6FD7
 
 | |
2MJF
 
 | | Solution structure of the complex between the yeast Rsa1 and Hit1 proteins | | Descriptor: | Protein HIT1, Ribosome assembly 1 protein | | Authors: | Quinternet, M, Roth, B, Back, R, Jacquemin, C, Manival, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction.
Nucleic Acids Res., 42, 2014
|
|
2MNJ
 
 | | NMR solution structure of the yeast Pih1 and Tah1 C-terminal domains complex | | Descriptor: | Protein interacting with Hsp90 1, TPR repeat-containing protein associated with Hsp90 | | Authors: | Quinternet, M, Jacquemin, C, Charpentier, B, Manival, X. | | Deposit date: | 2014-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D snoRNP Assembly.
J.Mol.Biol., 427, 2015
|
|
2M3F
 
 | | NMR structure of Rsa1p238-259 from S. Cerevisiae | | Descriptor: | Ribosome assembly 1 protein | | Authors: | Quinternet, M, Manival, X. | | Deposit date: | 2013-01-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly.
Nucleic Acids Res., 42, 2014
|
|
2GD1
 
 | |
