1B95
 
 | |
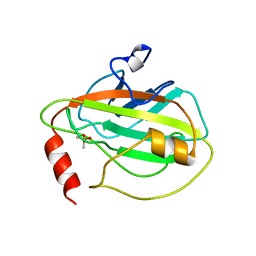
1BD9
 
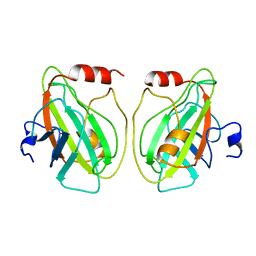 | |
1BEH
 
 | | HUMAN PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN IN COMPLEX WITH CACODYLATE | | Descriptor: | CACODYLATE ION, PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN | | Authors: | Banfield, M.J, Barker, J.J, Perry, A, Brady, R.L. | | Deposit date: | 1998-05-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction.
Structure, 6, 1998
|
|
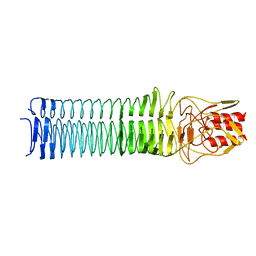
3PR7
 
 | |
1CDC
 
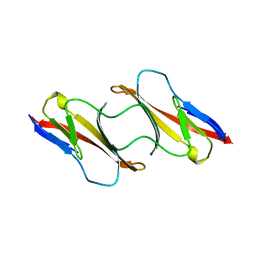 | | CD2, N-TERMINAL DOMAIN (1-99), TRUNCATED FORM | | Descriptor: | CD2 | | Authors: | Murray, A.J, Barclay, A.N, Brady, R.L. | | Deposit date: | 1995-05-23 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | One sequence, two folds: a metastable structure of CD2.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3R3K
 
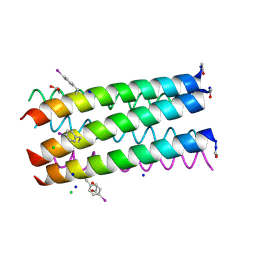 | | Crystal structure of a parallel 6-helix coiled coil | | Descriptor: | 1,2-ETHANEDIOL, CChex-Phi22 helix, CHLORIDE ION, ... | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.2009 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R46
 
 | | Crystal structure of a parallel 6-helix coiled coil CC-hex-D24 | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R4A
 
 | |
3R48
 
 | | Crystal structure of a hetero-hexamer coiled coil | | Descriptor: | GLYCEROL, coiled coil helix W22-L24H, coiled coil helix Y15-L24D | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0011 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R4H
 
 | |
3RA3
 
 | | Crystal structure of a section of a de novo design gigaDalton protein fibre | | Descriptor: | SODIUM ION, p1c, p2f | | Authors: | Zaccai, N.R, Sharp, T.H, Bruning, M, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cryo-transmission electron microscopy structure of a gigadalton peptide fiber of de novo design
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R8Q
 
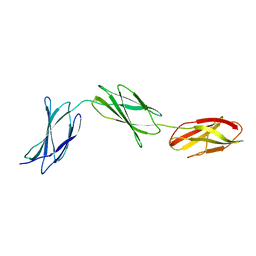 | |
3R9B
 
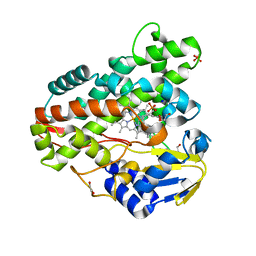 | | Crystal structure of Mycobacterium smegmatis CYP164A2 in ligand free state | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 164A2, DODECANE, ... | | Authors: | Agnew, C.R.J, Warrilow, A.G.S, Kelly, S.L, Brady, R.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | An enlarged, adaptable active site in CYP164 family P450 enzymes, the sole P450 in Mycobacterium leprae.
Antimicrob.Agents Chemother., 56, 2012
|
|
3R47
 
 | | Crystal structure of a 6-helix coiled coil CC-hex-H24 | | Descriptor: | BROMIDE ION, coiled coil helix L24H | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5002 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R9C
 
 | | Crystal structure of Mycobacterium smegmatis CYP164A2 with Econazole bound | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, Cytochrome P450 164A2, ... | | Authors: | Agnew, C.R.J, Kelly, S.L, Brady, R.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | An enlarged, adaptable active site in CYP164 family P450 enzymes, the sole P450 in Mycobacterium leprae.
Antimicrob.Agents Chemother., 56, 2012
|
|
1HE7
 
 | | Human Nerve growth factor receptor TrkA | | Descriptor: | GLYCEROL, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Banfield, M, Robertson, A, Allen, S, Dando, J, Tyler, S, Bennett, G, Brain, S, Mason, G, Holden, P, Clarke, A, Naylor, R, Wilcock, G, Brady, R, Dawbarn, D. | | Deposit date: | 2000-11-20 | | Release date: | 2001-04-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Structure of the Nerve Growth Factor Binding Site on Trka.
Biochem.Biophys.Res.Commun., 282, 2001
|
|
1LDG
 
 | | PLASMODIUM FALCIPARUM L-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C, Banfield, M, Barker, J, Higham, C, Moreton, K, Turgut-Balik, D, Brady, L, Holbrook, J.J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design.
Nat.Struct.Biol., 3, 1996
|
|
8A09
 
 | | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4 | | Descriptor: | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(QgLaId)4, GLYCEROL | | Authors: | Martin, F.J.O, Rhys, G.G, Dawson, W.M, Woolfson, D.N. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
6YB1
 
 | | Crystal structure of an antiparallel octameric transmembrane coiled coil K2-CCTM-VbIc | | Descriptor: | GLYCINE, K2-CCTM-VbIc, NONAETHYLENE GLYCOL | | Authors: | Kratochvil, H.T, Liu, L, Scott, A.J, Woolfson, D.N, DeGrado, W.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Constructing ion channels from water-soluble alpha-helical barrels.
Nat.Chem., 13, 2021
|
|
3H9E
 
 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
1R8H
 
 | | Comparison of the structure and DNA binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus | | Descriptor: | PHOSPHATE ION, Regulatory protein E2 | | Authors: | Dell, G, Wilkinson, K.W, Tranter, R, Parish, J, Brady, R.L, Gaston, K. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus.
J.Mol.Biol., 334, 2003
|
|
3PFW
 
 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD, a binary form | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
2BEY
 
 | |
