7GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-((N-ACETYL)-L-LEUCYLAMIDO)-4-PHENYL-BUTAN-2-ONE(N-ACETYL-L-LEUCYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
6GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
3GCH
 
 | |
4GCH
 
 | |
5GCH
 
 | |
7OP0
 
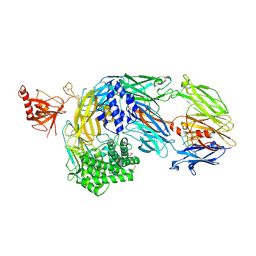 | | Crystal structure of complement C5 in complex with chemically synthesized K92 knob domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5 alpha chain, ... | | Authors: | Macpherson, A, van der Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Chemical Synthesis of Knob Domain Antibody Fragments.
Acs Chem.Biol., 16, 2021
|
|
3NS7
 
 | |
7P9J
 
 | |
7QAZ
 
 | |
8BAO
 
 | |
8BGJ
 
 | |
7NQD
 
 | |
7NQF
 
 | |
7NQE
 
 | |
6SA1
 
 | |
6SA0
 
 | |
