3TJ4
 
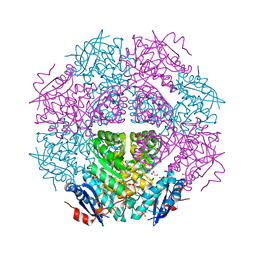 | | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg
to be published
|
|
4DXK
 
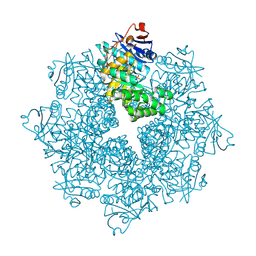 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme family protein, PHOSPHATE ION, SODIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate
to be published
|
|
4DN1
 
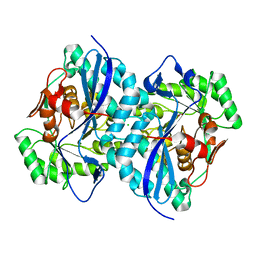 | | Crystal structure of an ENOLASE (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isomerase/lactonizing enzyme, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Sojitra, S, Al Obaidi, N.F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate
to be published
|
|
4DX3
 
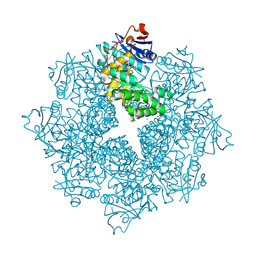 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase / muconate lactonizing enzyme family protein | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
to be published
|
|
4GGB
 
 | | Crystal structure of a proposed galactarolactone cycloisomerase from agrobacterium tumefaciens, TARGET EFI-500704, WITH BOUND CA, DISORDERED LOOPS | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4HP8
 
 | | Crystal structure of a putative 2-deoxy-d-gluconate 3-dehydrogenase from Agrobacterium Tumefaciens (target EFI-506435) with bound NADP | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vetting, M.W, Bouvier, J.T, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-23 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative 2-deoxy-d-gluconate 3-dehydrogenase from Agrobacterium Tumefaciens (target EFI-506435) with bound NADP
To be Published
|
|
4HPN
 
 | | Crystal structure of a proposed galactarolactone cycloisomerase from Agrobacterium Tumefaciens, target EFI-500704, with bound Ca, ordered loops | | Descriptor: | CALCIUM ION, IMIDAZOLE, NICKEL (II) ION, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4IBO
 
 | | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446) | | Descriptor: | CHLORIDE ION, Gluconate dehydrogenase, MAGNESIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446)
To be Published
|
|
4GFI
 
 | | Crystal structure of EFI-502318, an enolase family member from Agrobacterium tumefaciens with homology to dipeptide epimerases (bound sodium, L-Ala-L-Glu with ordered loop) | | Descriptor: | ALANINE, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EFI-502318, an enolase family member from Agrobacterium tumefaciens with homology to dipeptide epimerases (bound sodium, l-ala-l-glu with ordered loop)
To be Published
|
|
4H19
 
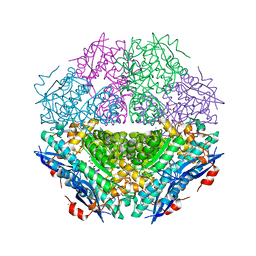 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop | | Descriptor: | (2R,3R,4R)-N,2,3,4,5-pentakis(oxidanyl)pentanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop
To be Published
|
|
4JN8
 
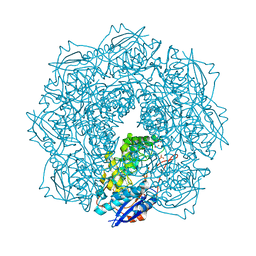 | | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ENOLASE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site
To be Published
|
|
4JN7
 
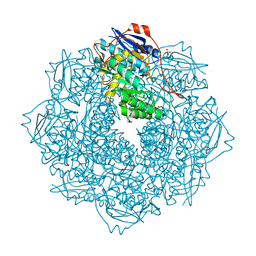 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4MUP
 
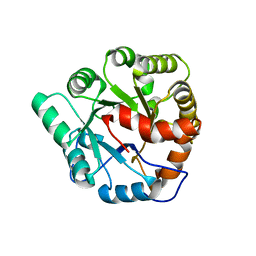 | | Crystal structure of Agrobacterium tumefaciens ATU3138 (EFI target 505157), apo structure | | Descriptor: | AMIDOHYDROLASE | | Authors: | Vetting, M.W, Bouvier, J.T, Groninger-Poe, F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Agrobacterium tumefaciens ATU3138 (EFI target 505157), apo structure
To be Published
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-25 | | Release date: | 2014-03-05 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVS
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFUROSPIRILLUM DELEYIANUM DSM 6946 (Sdel_0447), TARGET EFI-510309, WITH BOUND SUCCINATE | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OA4
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SHEWANELLA LOIHICA PV-4 (Shew_1446), TARGET EFI-510273, WITH BOUND SUCCINATE | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-05 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVT
 
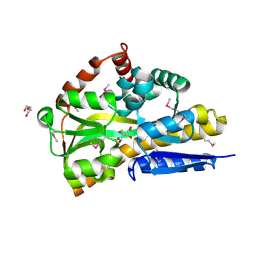 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTERIUM ANTHROPI (Oant_3902), TARGET EFI-510153, WITH BOUND L-FUCONATE | | Descriptor: | 6-deoxy-L-galactonic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OAN
 
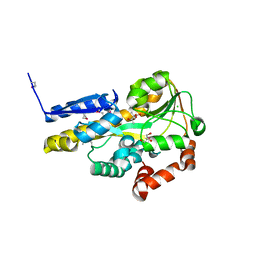 | | Crystal structure of a TRAP periplasmic solute binding protein from rhodopseudomonas palustris HaA2 (RPB_2686), TARGET EFI-510221, with density modeled as (S)-2-hydroxy-2-methyl-3-oxobutanoate ((S)-2-Acetolactate) | | Descriptor: | (2S)-2-hydroxy-2-methyl-3-oxobutanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-05 | | Release date: | 2014-01-22 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVR
 
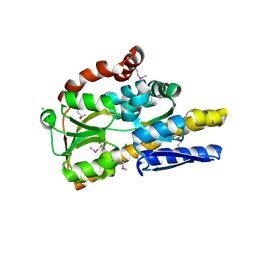 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM XANTHOBACTER AUTOTROPHICUS PY2, TARGET EFI-510329, WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P3L
 
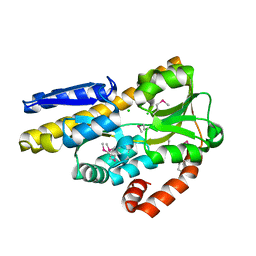 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CHROMOHALOBACTER SALEXIGENS DSM 3043 (Csal_2479), TARGET EFI-510085, WITH BOUND GLUCURONATE, SPG P6122 | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-09 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
